1G1U
 
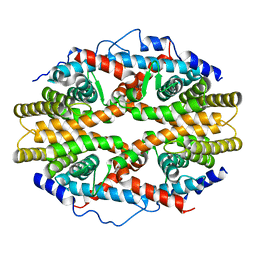 | | THE 2.5 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE RXRALPHA LIGAND BINDING DOMAIN IN TETRAMER IN THE ABSENCE OF LIGAND | | Descriptor: | RETINOIC ACID RECEPTOR RXR-ALPHA | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Wisely, G.B, Milburn, M.V, Xu, H.E. | | Deposit date: | 2000-10-13 | | Release date: | 2001-04-25 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix.
Genes Dev., 14, 2000
|
|
1G5Y
 
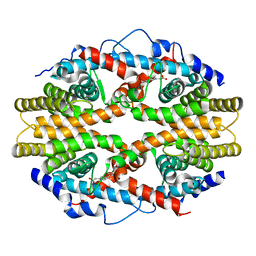 | | THE 2.0 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE RXRALPHA LIGAND BINDING DOMAIN TETRAMER IN THE PRESENCE OF A NON-ACTIVATING RETINOIC ACID ISOMER. | | Descriptor: | RETINOIC ACID, RETINOIC ACID RECEPTOR RXR-ALPHA | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Wisely, G.B, Milburn, M.V, Xu, H.E. | | Deposit date: | 2000-11-02 | | Release date: | 2001-05-02 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix.
Genes Dev., 14, 2000
|
|
6IN9
 
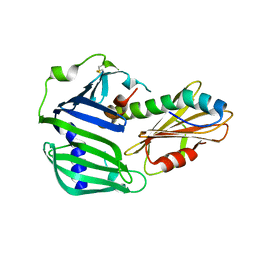 | | Crystal structure of MucB in complex with MucA(peri) | | Descriptor: | Sigma factor AlgU negative regulatory protein, Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
1GLO
 
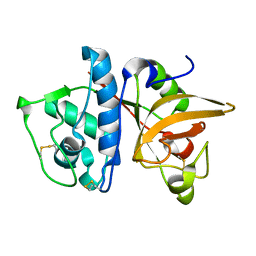 | | Crystal Structure of Cys25Ser mutant of human cathepsin S | | Descriptor: | CATHEPSIN S | | Authors: | Turkenburg, J.P, Lamers, M.B.A.C, Brzozowski, A.M, Wright, L.M, Hubbard, R.E, Sturt, S.L, Williams, D.H. | | Deposit date: | 2001-08-31 | | Release date: | 2002-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a Cys25->Ser Mutant of Human Cathepsin Cathepsin S
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6IKK
 
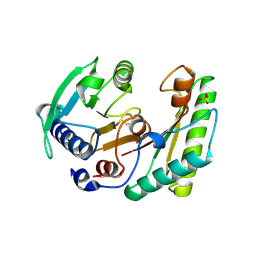 | |
1JS1
 
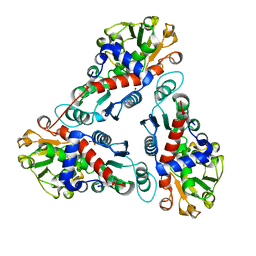 | | Crystal Structure of a new transcarbamylase from the anaerobic bacterium Bacteroides fragilis at 2.0 A resolution | | Descriptor: | PHOSPHATE ION, Transcarbamylase | | Authors: | Shi, D, Gallegos, R, DePonte III, J, Morizono, H, Yu, X, Allewell, N.M, Malamy, M, Tuchman, M. | | Deposit date: | 2001-08-15 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a transcarbamylase-like protein from the anaerobic bacterium Bacteroides fragilis at 2.0 A resolution.
J.Mol.Biol., 320, 2002
|
|
6INC
 
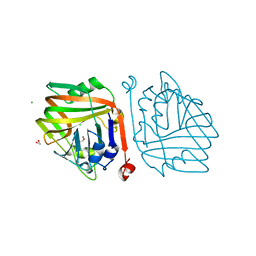 | | Crystal structure of an acetolactate decarboxylase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Alpha-acetolactate decarboxylase, CHLORIDE ION, ... | | Authors: | Wu, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural characterization of an acetolactate decarboxylase from Klebsiella pneumoniae
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6JC4
 
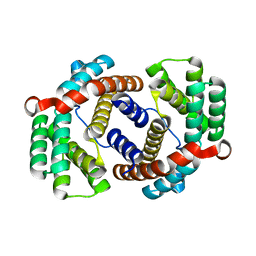 | |
4F76
 
 | |
4F75
 
 | |
1NRL
 
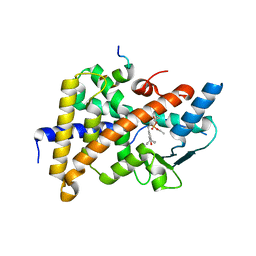 | | Crystal Structure of the human PXR-LBD in complex with an SRC-1 coactivator peptide and SR12813 | | Descriptor: | Nuclear Receptor Coactivator 1 isoform 3, Orphan nuclear receptor PXR, [2-(3,5-DI-TERT-BUTYL-4-HYDROXY-PHENYL)-1-(DIETHOXY-PHOSPHORYL)-VINYL]-PHOSPHONIC ACID DIETHLYL ESTER | | Authors: | Watkins, R.E, Davis-Searles, P.R, Lambert, M.H, Redinbo, M.R. | | Deposit date: | 2003-01-25 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor
J.Mol.Biol., 331, 2003
|
|
6INB
 
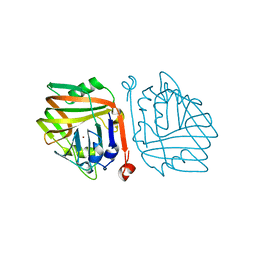 | |
6IKI
 
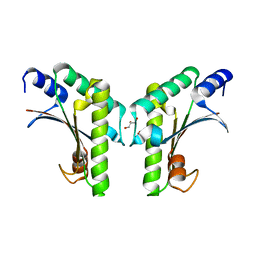 | | Crystal structure of YfiB(W55L) | | Descriptor: | GLYCEROL, SULFATE ION, YfiB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural analysis of activating mutants of YfiB from Pseudomonas aeruginosa PAO1.
Biochem. Biophys. Res. Commun., 506, 2018
|
|
5WQO
 
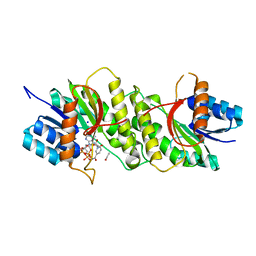 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition I) | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable dehydrogenase, ... | | Authors: | Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2016-11-27 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
4F74
 
 | |
5WQM
 
 | |
4F73
 
 | |
1K7L
 
 | | The 2.5 Angstrom resolution crystal structure of the human PPARalpha ligand binding domain bound with GW409544 and a co-activator peptide. | | Descriptor: | 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor alpha, YTTRIUM (III) ION, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-19 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3GT8
 
 | | Crystal structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jura, N, Endres, N.F, Engel, K, Deindl, S, Das, R, Lamers, M.H, Wemmer, D.E, Zhang, X, Kuriyan, J. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment.
Cell(Cambridge,Mass.), 137, 2009
|
|
8S7Z
 
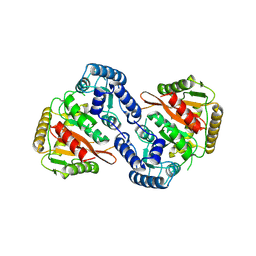 | |
8HJB
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA with coenzyme A | | Descriptor: | COENZYME A, TetR family transcriptional regulator | | Authors: | Liang, H, Bartlam, M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
5FDL
 
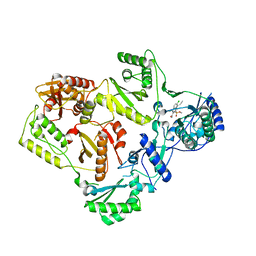 | | Crystal Structure of K103N/Y181C Mutant HIV-1 Reverse Transcriptase (RT) in Complex with IDX899 | | Descriptor: | P51 Reverse transcriptase, P66 Reverse transcriptase, methyl (R)-(2-carbamoyl-5-chloro-1H-indol-3-yl)[3-(2-cyanoethyl)-5-methylphenyl]phosphinate | | Authors: | Dousson, C.B, Alexandre, F.-R, Convard, T, Fisher, M, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2015-12-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of the Aryl-phospho-indole IDX899, a Highly Potent Anti-HIV Non-nucleoside Reverse Transcriptase Inhibitor.
J.Med.Chem., 59, 2016
|
|
1P9E
 
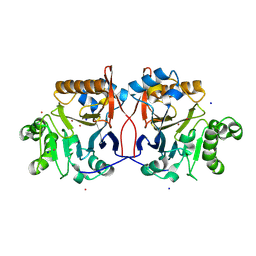 | | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3 | | Descriptor: | CADMIUM ION, Methyl Parathion Hydrolase, POTASSIUM ION, ... | | Authors: | Dong, Y, Sun, L, Bartlam, M, Rao, Z, Zhang, X. | | Deposit date: | 2003-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3
To be Published
|
|
4FTH
 
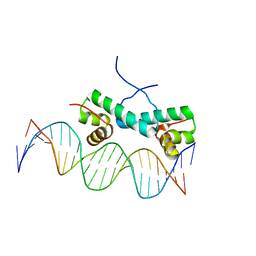 | | Crystal Structure of NtrC4 DNA-binding domain bound to double-stranded DNA | | Descriptor: | 5'-D(*AP*CP*TP*TP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*AP*AP*AP*TP*GP*CP*AP*T)-3', 5'-D(P*GP*AP*TP*GP*CP*AP*TP*TP*TP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*AP*A)-3', Transcriptional regulator (NtrC family) | | Authors: | Vidangos, N.K, Heideker, J, Lyubimov, A.Y, Lamers, M, Huo, Y, Pelton, J.G, Ton, J, Gralla, J.D, Kuriyan, J, Berger, J.M, Wemmer, D.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | DNA Recognition by a sigma (54) Transcriptional Activator from Aquifex aeolicus.
J.Mol.Biol., 426, 2014
|
|
1K74
 
 | | The 2.3 Angstrom resolution crystal structure of the heterodimer of the human PPARgamma and RXRalpha ligand binding domains respectively bound with GW409544 and 9-cis retinoic acid and co-activator peptides. | | Descriptor: | (9cis)-retinoic acid, 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor gamma, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-18 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
