8T8T
 
 | |
4WKM
 
 | | AmpR effector binding domain from Citrobacter freundii bound to UDP-MurNAc-pentapeptide | | Descriptor: | ALA-FGA-API-DAL-DAL, GLYCEROL, LysR family transcriptional regulator, ... | | Authors: | Vadlamani, G, Reeve, T.M, Mark, B.L. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The beta-Lactamase Gene Regulator AmpR Is a Tetramer That Recognizes and Binds the d-Ala-d-Ala Motif of Its Repressor UDP-N-acetylmuramic Acid (MurNAc)-pentapeptide.
J.Biol.Chem., 290, 2015
|
|
5K69
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T224 | | Descriptor: | (2~{S},3~{R},4~{R})-4-(1~{H}-indol-3-ylsulfanyl)-3-methyl-2-[(2~{S},3~{S})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, GLYCEROL, L,D-transpeptidase 2 | | Authors: | Lamichhane, G, Ginell, S.L, Kumar, P. | | Deposit date: | 2016-05-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5UTQ
 
 | |
5UTP
 
 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to N-ethylbutyryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, N-[(2Z,3R,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)-2-{[(phenylcarbamoyl)oxy]imino}tetrahydro-2H-pyran-3-yl]-2-ethylbutanamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2017-02-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational flexibility of the glycosidase NagZ allows it to bind structurally diverse inhibitors to suppress beta-lactam antibiotic resistance.
Protein Sci., 26, 2017
|
|
5UTR
 
 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-butyryl-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]butanamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2017-02-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational flexibility of the glycosidase NagZ allows it to bind structurally diverse inhibitors to suppress beta-lactam antibiotic resistance.
Protein Sci., 26, 2017
|
|
4MSS
 
 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-acetamido-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase 1, GLYCEROL, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]acetamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2013-09-18 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective trihydroxyazepane NagZ inhibitors increase sensitivity of Pseudomonas aeruginosa to beta-lactams.
Chem.Commun.(Camb.), 49, 2013
|
|
7MMY
 
 | |
7MPY
 
 | |
5FCZ
 
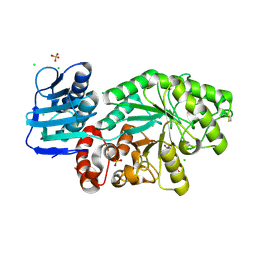 | |
5FD0
 
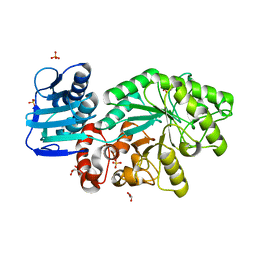 | |
8TWH
 
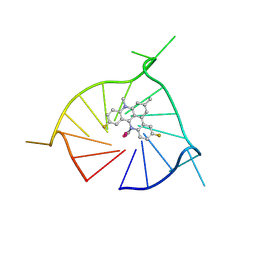 | | Crystal structure of (GGGTT)3GGG G-quadruplex in complex with small molecule ligand RHPS4 | | Descriptor: | (GGGTT)3GGG DNA, 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, POTASSIUM ION | | Authors: | Yatsunyk, L.A, Martin, K.N, Lam, G. | | Deposit date: | 2023-08-21 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of (GGGTT)3GGG G-quadruplex in complex with small molecule ligand RHPS4
To be published
|
|
5DUJ
 
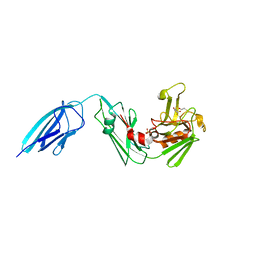 | | Crystal structure of ldtMt2 in complex with Faropenem adduct | | Descriptor: | (2R,5R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydro-1,3-thiazole-4-carboxylic acid, BETA-MERCAPTOETHANOL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5DU7
 
 | |
5E1I
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T210 | | Descriptor: | (2S,3R,4R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-(methylsulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, GLYCEROL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-29 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5DZJ
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T206 in conformation A | | Descriptor: | (2~{R},3~{R},4~{R})-4-methyl-3-(2-oxidanylidene-2-propoxy-ethyl)sulfanyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, L,D-transpeptidase 2 | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
3TX4
 
 | | Crystal Structure of Mutant (C354A) M. tuberculosis LD-transpeptidase type 2 | | Descriptor: | Mycobacterium Tuberculosis LD-transpeptidase type 2 | | Authors: | Erdemli, S, Bianchet, M.A, Gupta, R, Lamichhane, G, Amzel, L.M. | | Deposit date: | 2011-09-22 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Targeting the Cell Wall of Mycobacterium tuberculosis: Structure and Mechanism of L,D-Transpeptidase 2.
Structure, 20, 2012
|
|
6Q42
 
 | | Crystal Structure of Human Pancreatic Phospholipase A2 | | Descriptor: | CHLORIDE ION, Phospholipase A2 | | Authors: | Saul, F, Haouz, A, Lambeau, G, Theze, J. | | Deposit date: | 2018-12-05 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PLA2G1B is involved in CD4 anergy and CD4 lymphopenia in HIV-infected patients.
J.Clin.Invest., 130, 2020
|
|
5I6C
 
 | | The structure of the eukaryotic purine/H+ symporter, UapA, in complex with Xanthine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Uric acid-xanthine permease, XANTHINE | | Authors: | Alguel, Y, Amillis, S, Leung, J, Lambrinidis, G, Capaldi, S, Scull, N.J, Craven, G, Iwata, S, Armstrong, A, Mikros, E, Diallinas, G, Cameron, A.D, Byrne, B. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-27 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of eukaryotic purine/H(+) symporter UapA suggests a role for homodimerization in transport activity.
Nat Commun, 7, 2016
|
|
6MK6
 
 | |
4Z7A
 
 | | Structural and biochemical characterization of a non-functionally redundant M. tuberculosis (3,3) L,D-Transpeptidase, LdtMt5. | | Descriptor: | ACETYL GROUP, DI(HYDROXYETHYL)ETHER, Mycobacterium tuberculosis (3,3)L,D-Transpeptidase type 5, ... | | Authors: | Basta, L, Ghosh, A, Pan, Y, Jakoncic, J, Lloyd, E, Townsend, G, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-04-06 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Loss of a Functionally and Structurally Distinct ld-Transpeptidase, LdtMt5, Compromises Cell Wall Integrity in Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
4GV1
 
 | | PKB alpha in complex with AZD5363 | | Descriptor: | 4-amino-N-[(1S)-1-(4-chlorophenyl)-3-hydroxypropyl]-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamide, GLYCEROL, RAC-alpha serine/threonine-protein kinase | | Authors: | Addie, M, Ballard, P, Bird, G, Buttar, D, Currie, G, Davies, B, Debreczeni, J, Dry, H, Dudley, P, Greenwood, R, Hatter, G, Jestel, A, Johnson, P.D, Kettle, J.G, Lane, C, Lamont, G, Leach, A, Luke, R.W.A, Ogilvie, D, Page, K, Pass, M, Steinbacher, S, Steuber, H, Pearson, S, Ruston, L. | | Deposit date: | 2012-08-30 | | Release date: | 2013-02-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of 4-Amino-N-[(1S)-1-(4-chlorophenyl)-3-hydroxypropyl]-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamide (AZD5363), an Orally Bioavailable, Potent Inhibitor of Akt Kinases.
J.Med.Chem., 56, 2013
|
|
5UWV
 
 | | Crystal structure of Mycobacterium abscessus L,D-transpeptidase 2 | | Descriptor: | L,D-TRANSPEPTIDASE 2 | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2017-02-21 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Mycobacterium abscessus l,d-Transpeptidases Are Susceptible to Inactivation by Carbapenems and Cephalosporins but Not Penicillins.
Antimicrob. Agents Chemother., 61, 2017
|
|
4ZFQ
 
 | | Structure of M. tuberculosis (3,3) L,D-Transpeptidase, LdtMt5. (Meropenen-adduct form) | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, DI(HYDROXYETHYL)ETHER, L,D-transpeptidase 5 | | Authors: | Basta, L, Ghosh, A, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-04-21 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Loss of a Functionally and Structurally Distinct ld-Transpeptidase, LdtMt5, Compromises Cell Wall Integrity in Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
4O9R
 
 | | Human Smoothened Receptor structure in complex with cyclopamine | | Descriptor: | Cyclopamine, Smoothened homolog/Soluble cytochrome b562 chimeric protein | | Authors: | Wang, C, Weierstall, U, James, D, White, T.A, Wang, D, Liu, W, Spence, J.C.H, Doak, R.B, Nelson, G, Fromme, P, Fromme, R, Grotjohann, I, Kupitz, C, Zatsepin, N.A, Liu, H, Basu, S, Wacker, D, Han, G.W, Katritch, V, Boutet, S, Messerschmidt, M, Willams, G.J, Koglin, J.E, Seibert, M.M, Klinker, M, Gati, C, Shoeman, R.L, Barty, A, Chapman, H.N, Kirian, R.A, Beyerlein, K.R, Stevens, R.C, Li, D, Shah, S.T.A, Howe, N, Caffrey, M, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography.
Nat Commun, 5, 2014
|
|
