2NNY
 
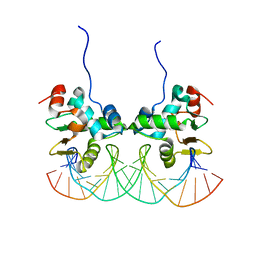 | | Crystal structure of the Ets1 dimer DNA complex. | | Descriptor: | 5'-D(*A*CP*TP*CP*CP*AP*GP*GP*AP*AP*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*TP*CP*T)-3', 5'-D(*T*AP*GP*AP*CP*AP*GP*GP*AP*AP*GP*CP*AP*CP*TP*TP*CP*CP*TP*GP*GP*AP*G)-3', C-ets-1 protein | | Authors: | Lamber, E.P, Kachalova, G.S, Wilmanns, M. | | Deposit date: | 2006-10-24 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Regulation of the transcription factor Ets-1 by DNA-mediated homo-dimerization.
Embo J., 27, 2008
|
|
4PWA
 
 | |
2VNV
 
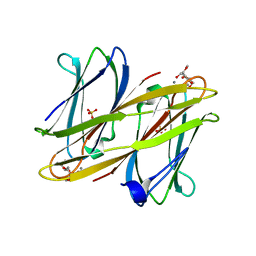 | | Crystal structure of BclA lectin from burkholderia cenocepacia in complex with alpha-methyl-mannoside at 1.7 Angstrom resolution | | Descriptor: | BCLA, CALCIUM ION, SULFATE ION, ... | | Authors: | Lameignere, E, Malinovska, L, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Mannose Recognition by a Lectin from Opportunistic Bacteria Burkholderia Cenocepacia
Biochem.J., 411, 2008
|
|
2VFC
 
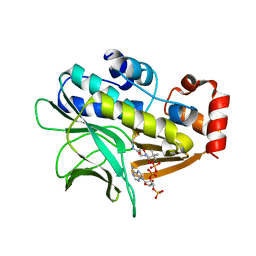 | | The structure of Mycobacterium marinum arylamine N-acetyltransferase in complex with CoA | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE, COENZYME A | | Authors: | Fullam, E, Westwood, I.M, Anderton, M.C, Lowe, E.D, Sim, E, Noble, M.E.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Divergence of Cofactor Recognition Across Evolution: Coenzyme a Binding in a Prokaryotic Arylamine N-Acetyltransferase.
J.Mol.Biol., 375, 2008
|
|
2VFB
 
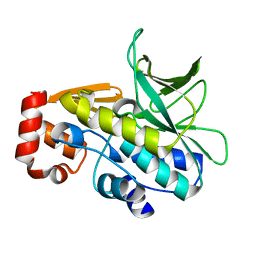 | | The structure of Mycobacterium marinum arylamine N-acetyltransferase | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Fullam, E, Westwood, I.M, Anderton, M.C, Lowe, E.D, Sim, E, Noble, M.E.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Divergence of Cofactor Recognition Across Evolution: Coenzyme a Binding in a Prokaryotic Arylamine N-Acetyltransferase.
J.Mol.Biol., 375, 2008
|
|
2WR9
 
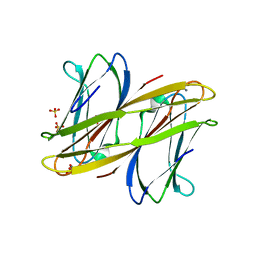 | | CRYSTAL STRUCTURE OF BURKHOLDERIA CENOCEPACIA LECTIN (BCLA) COMPLEXED WITH AMAN1-3MAN DISACCHARIDE | | Descriptor: | CALCIUM ION, LECTIN, SULFATE ION, ... | | Authors: | Lameignere, E, Shiao, T.C, Roy, R, Wimmerova, M, Dubreuil, F, Varrot, A, Imberty, A. | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of the Affinity for Oligomannosides and Analogs Displayed by Bc2L-A, a Burkholderia Cenocepacia Soluble Lectin.
Glycobiology, 20, 2010
|
|
2WRA
 
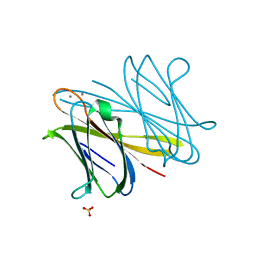 | | BclA lectin from Burkholderia cenocepacia complexed with aMan1(aMan1- 6)-3Man trisaccharide | | Descriptor: | CALCIUM ION, LECTIN, SULFATE ION, ... | | Authors: | Lameignere, E, Shiao, T.C, Roy, R, Wimmerova, M, Dubreuil, F, Varrot, A, Imberty, A. | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Basis of the Affinity for Oligomannosides and Analogs Displayed by Bc2L-A, a Burkholderia Cenocepacia Soluble Lectin.
Glycobiology, 20, 2010
|
|
3TU6
 
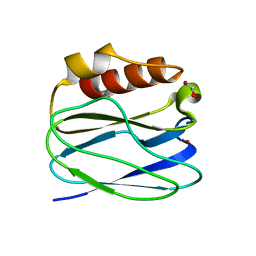 | | The Structure of a Pseudoazurin From Sinorhizobium meliltoi | | Descriptor: | COPPER (II) ION, GLYCEROL, Pseudoazurin (Blue copper protein) | | Authors: | Laming, E.M, McGrath, A.P, Guss, J.M, Maher, M.J. | | Deposit date: | 2011-09-16 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray crystal structure of a pseudoazurin from Sinorhizobium meliloti.
J.Inorg.Biochem., 115, 2012
|
|
6W8R
 
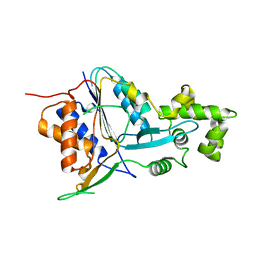 | | Crystal structure of metacaspase 4 C139A from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6W8S
 
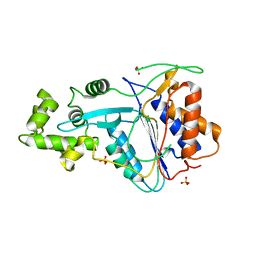 | | Crystal structure of metacaspase 4 from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6W8T
 
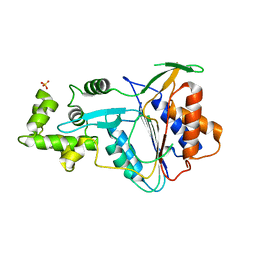 | | Crystal structure of metacaspase 4 from Arabidopsis (microcrystals treated with calcium) | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
7APE
 
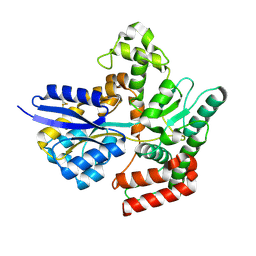 | | Crystal structure of LpqY from Mycobacterium thermoresistible in complex with trehalose | | Descriptor: | Lipoprotein (Sugar-binding) lpqY, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Furze, C.M, Guy, C.M, Angula, J, Cameron, A.D, Fullam, E. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of trehalose recognition by the mycobacterial LpqY-SugABC transporter.
J.Biol.Chem., 296, 2021
|
|
6FV3
 
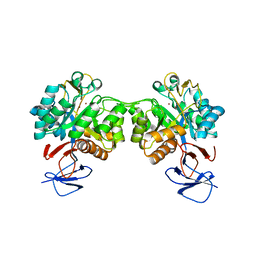 | | Crystal structure of N-acetyl-D-glucosamine-6-phosphate deacetylase from Mycobacterium smegmatis. | | Descriptor: | N-acetylglucosamine-6-phosphate deacetylase, ZINC ION | | Authors: | Ahangar, M.S, Furze, C.M, Guy, C.S, Cooper, C, Maskew, K.S, Graham, B, Cameron, A.D, Fullam, E. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and functional determination of homologs of theMycobacterium tuberculosis N-acetylglucosamine-6-phosphate deacetylase (NagA).
J. Biol. Chem., 293, 2018
|
|
6R1B
 
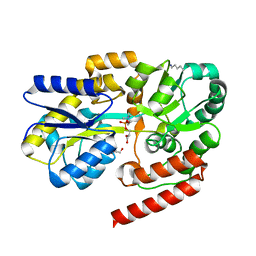 | | Crystal structure of UgpB from Mycobacterium tuberculosis in complex with glycerophosphocholine | | Descriptor: | 2-(((R)-2,3-DIHYDROXYPROPYL)PHOSPHORYLOXY)-N,N,N-TRIMETHYLETHANAMINIUM, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fenn, J, Nepravishta, R, Guy, C.S, Harrison, J, Angulo, J, Cameron, A.D, Fullam, E. | | Deposit date: | 2019-03-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27000213 Å) | | Cite: | Structural Basis of Glycerophosphodiester Recognition by theMycobacterium tuberculosisSubstrate-Binding Protein UgpB.
Acs Chem.Biol., 14, 2019
|
|
6FV4
 
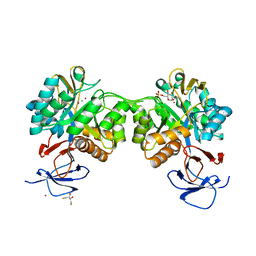 | | The structure of N-acetyl-D-glucosamine-6-phosphate deacetylase D267A mutant from Mycobacterium smegmatis in complex with N-acetyl-D-glucosamine-6-phosphate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, CADMIUM ION, ... | | Authors: | Ahangar, M.S, Furze, C.M, Guy, C.S, Cooper, C, Maskew, K.S, Graham, B, Cameron, A.D, Fullam, E. | | Deposit date: | 2018-03-01 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structural and functional determination of homologs of theMycobacterium tuberculosis N-acetylglucosamine-6-phosphate deacetylase (NagA).
J. Biol. Chem., 293, 2018
|
|
5K2X
 
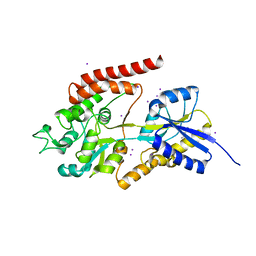 | | Crystal structure of M. tuberculosis UspC (tetragonal crystal form) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, IODIDE ION, ... | | Authors: | Futterer, K, Fullam, E, Besra, G.S. | | Deposit date: | 2016-05-19 | | Release date: | 2016-06-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional analysis of the solute-binding protein UspC from Mycobacterium tuberculosis that is specific for amino sugars.
Open Biology, 6, 2016
|
|
5K2Y
 
 | |
8CR6
 
 | | mouse Interleukin-12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 subunit alpha, Interleukin-12 subunit beta, ... | | Authors: | Merceron, R, Felix, J, Lambert, E, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5LJP
 
 | |
1EGU
 
 | | CRYSTAL STRUCTURE OF STREPTOCOCCUS PNEUMONIAE HYALURONATE LYASE AT 1.56 A RESOLUTION | | Descriptor: | HYALURONATE LYASE, SULFATE ION | | Authors: | Li, S, Kelly, S.J, Lamani, E, Ferraroni, M, Jedrzejas, M.J. | | Deposit date: | 2000-02-16 | | Release date: | 2001-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase.
EMBO J., 19, 2000
|
|
4B55
 
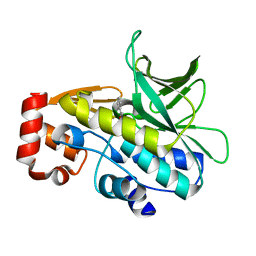 | | Crystal Structure of the Covalent Adduct Formed between Mycobacterium marinum Aryalamine N-acetyltransferase and Phenyl vinyl ketone a derivative of Piperidinols | | Descriptor: | 3-hydroxy-1-phenylpropan-1-one, ARYLAMINE N-ACETYLTRANSFERASE NAT | | Authors: | Abuhammad, A, Fullam, E, Lowe, E.D, Staunton, D, Kawamura, A, Westwood, I.M, Bhakta, S, Garner, A.C, Wilson, D.L, Seden, P.T, Davies, S.G, Russell, A.J, Garman, E.F, Sim, E. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Piperidinols that Show Anti-Tubercular Activity as Inhibitors of Arylamine N-Acetyltransferase: An Essential Enzyme for Mycobacterial Survival Inside Macrophages.
Plos One, 7, 2012
|
|
4AUT
 
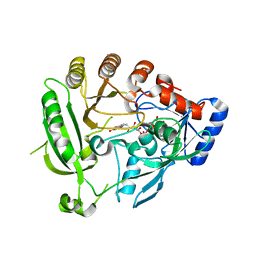 | | Crystal structure of the tuberculosis drug target Decaprenyl- Phosphoryl-beta-D-Ribofuranose-2-oxidoreductase (DprE1) from Mycobacterium smegmatis | | Descriptor: | DECAPRENYL-PHOSPHORYL-BETA-D-RIBOFURANOSE-2-OXIDOREDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Neres, J, Pojer, F, Molteni, E, Chiarelli, L.R, Dhar, N, Boy-Rottger, S, Buroni, S, Fullam, E, Degiacomi, G, Lucarelli, A, Read, R.J, Zanoni, G, Edmondson, D.E, De Rossi, E, Pasca, M, Riccardi, G, Mattevi, A, Dyson, P.J, Cole, S.T, Binda, C. | | Deposit date: | 2012-05-21 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Benzothiazinone-Mediated Killing of Mycobacterium Tuberculosis.
Sci. Transl. Med., 4, 2012
|
|
4AOC
 
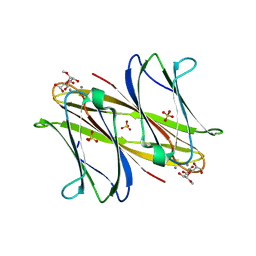 | | crystal structure of BC2L-A Lectin from Burkolderia cenocepacia in complex with methyl-heptoside | | Descriptor: | BC2L-A LECTIN, CALCIUM ION, SULFATE ION, ... | | Authors: | Marchetti, R, Malinovska, L, Lameignere, E, deCastro, C, Cioci, G, Kosma, P, Wimmerova, M, Molinaro, A, Imberty, A, Silipo, A. | | Deposit date: | 2012-03-26 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Burkholderia Cenocepacia Lectin a Binding to Heptoses from the Bacterial Lipopolysaccharide.
Glycobiology, 22, 2012
|
|
1XG4
 
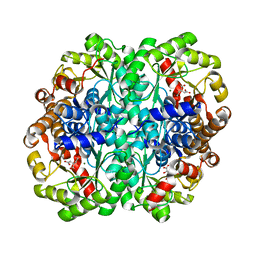 | | Crystal Structure of the C123S 2-Methylisocitrate Lyase Mutant from Escherichia coli in complex with the inhibitor isocitrate | | Descriptor: | ISOCITRIC ACID, MAGNESIUM ION, Probable methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Han, Y, Melamud, E, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of 2-Methylisocitrate Lyase in Complex with Product and with Isocitrate Inhibitor Provide Insight into Lyase Substrate Specificity, Catalysis and Evolution
Biochemistry, 44, 2005
|
|
1XG3
 
 | | Crystal structure of the C123S 2-methylisocitrate lyase mutant from Escherichia coli in complex with the reaction product, Mg(II)-pyruvate and succinate | | Descriptor: | MAGNESIUM ION, PYRUVIC ACID, Probable methylisocitrate lyase, ... | | Authors: | Liu, S, Lu, Z, Han, Y, Melamud, E, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of 2-Methylisocitrate Lyase in Complex with Product and with Isocitrate Inhibitor Provide Insight into Lyase Substrate Specificity, Catalysis and Evolution
Biochemistry, 44, 2005
|
|
