6GZH
 
 | | Crystal Structure of Human CDK9/cyclinT1 with A86 | | Descriptor: | Cyclin-T1, Cyclin-dependent kinase 9, GLYCEROL, ... | | Authors: | Ben-neriah, Y, Venkatachalam, A, Minzel, W, Fink, A, Snir-Alkalay, I, Vacca, J. | | Deposit date: | 2018-07-04 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Small Molecules Co-targeting CKI alpha and the Transcriptional Kinases CDK7/9 Control AML in Preclinical Models.
Cell, 175, 2018
|
|
6C4N
 
 | | Pseudopaline dehydrogenase (PaODH) - NADP+ bound | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pseudopaline dehydrogenase | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
5V0Q
 
 | |
5ZKV
 
 | | Solution structure of molten globule state of L94G mutant of horse cytochrome-c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Naiyer, A, Islam, A, Hassan, M.I, Sundd, M, Ahmad, F. | | Deposit date: | 2018-03-26 | | Release date: | 2019-05-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of molten globule state of L94G mutant of horse cytochrome-c
To Be Published
|
|
8B5T
 
 | |
8B5U
 
 | |
4KYP
 
 | | Beta-Scorpion Toxin folded in the periplasm of E.coli | | Descriptor: | Beta-insect excitatory toxin Bj-xtrIT, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | O'Reilly, A.O, Cole, A.R, Lopes, J.L, Lampert, A, Wallace, B.A. | | Deposit date: | 2013-05-29 | | Release date: | 2014-02-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chaperone-mediated native folding of a beta-scorpion toxin in the periplasm of Escherichia coli.
Biochim.Biophys.Acta, 1840, 2014
|
|
8BM3
 
 | | H207A mutant of E. coli PgpB, a PAP2 type phosphatidyl glycerol phosphate and C55-PP phosphatase, in complex with farnesyl pyrophosphate | | Descriptor: | FARNESYL DIPHOSPHATE, Phosphatidylglycerophosphatase B | | Authors: | Delbrassine, F, El Ghachi, M, Lambion, A, Herman, R, Kerff, F. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | H207A mutant of E. coli PgpB, a PAP2 type phosphatidyl glycerol phosphate and C55-PP phosphatase, in complex with farnesyl pyrophosphate
To Be Published
|
|
3TCO
 
 | | Crystallographic and spectroscopic characterization of Sulfolobus solfataricus TrxA1 provide insights into the determinants of thioredoxin fold stability | | Descriptor: | 1,2-ETHANEDIOL, Thioredoxin (TrxA-1) | | Authors: | Esposito, L, Ruggiero, A, Masullo, M, Ruocco, M.R, Lamberti, A, Arcari, P, Zagari, A, Vitagliano, L. | | Deposit date: | 2011-08-09 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and spectroscopic characterizations of Sulfolobus solfataricus TrxA1 provide insights into the determinants of thioredoxin fold stability.
J.Struct.Biol., 177, 2012
|
|
2WPE
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00073359) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
3H80
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213 | | Descriptor: | 1,2-ETHANEDIOL, Heat shock protein 83-1, MAGNESIUM ION, ... | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, Mackenzie, F, Fairlamb, A, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213
To be Published
|
|
3H5Z
 
 | | Crystal Structure of Leishmania major N-myristoyltransferase with bound myristoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, ... | | Authors: | Qiu, W, Hutchinson, A, Lin, Y.-H, Wernimont, A, Mackenzie, F, Ravichandran, M, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Fairlamb, A.H, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | N-myristoyltransferase inhibitors as new leads to treat sleeping sickness.
Nature, 464, 2010
|
|
3U67
 
 | | Crystal structure of the N-terminal domain of Hsp90 from Leishmania major(LmjF33.0312)in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 83-1, ... | | Authors: | Pizarro, J.C, Wernimont, A.K, Hutchinson, A, Mackenzie, F, Fairlamb, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-12 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
3Q5L
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin | | Descriptor: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-19-{[2-(pyrrolidin-1-yl)ethyl]amino}-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin.
To be Published
|
|
3Q5J
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin | | Descriptor: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-19-{[3-(dimethylamino)propyl]amino}-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin.
To be Published
|
|
7P3I
 
 | | Crystal structure of human CD40/TNFRSF5 in complex with the anti-CD40 DARPin protein | | Descriptor: | Darpin, SODIUM ION, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Malvezzi, F, Mangold, S, Hospodarsch, T, Reichen, C, Iss, C, Lammens, A, Krapp, S, Domke, C. | | Deposit date: | 2021-07-07 | | Release date: | 2022-04-06 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Multispecific Anti-CD40 DARPin Construct Induces Tumor-Selective CD40 Activation and Tumor Regression.
Cancer Immunol Res, 10, 2022
|
|
2RKJ
 
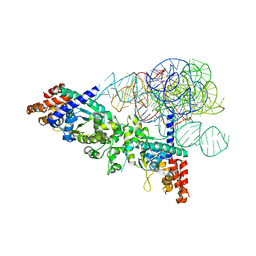 | | Cocrystal structure of a tyrosyl-tRNA synthetase splicing factor with a group I intron RNA | | Descriptor: | RNA (238-MER), RNA (5'-R(P*GP*CP*UP*U)-3'), Tyrosyl-tRNA synthetase | | Authors: | Paukstelis, P.J, Chen, J.-H, Chase, E, Lambowitz, A.M, Golden, B.L. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA.
Nature, 451, 2008
|
|
4M2A
 
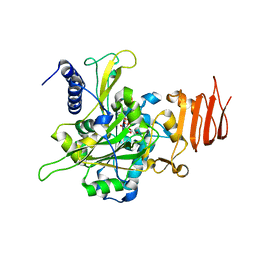 | | Crystal structure of the udp-glucose pyrophosphorylase from Leishmania major in the post-reactive state | | Descriptor: | MAGNESIUM ION, SULFATE ION, UDP-glucose pyrophosphorylase, ... | | Authors: | Fuehring, J, Routier, F.H, Lamerz, A.-C, Baruch, P, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2013-08-05 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Catalytic Mechanism and Allosteric Regulation of Udp-Glucose Pyrophosphorylase from Leishmania Major
ACS CATALYSIS, 3, 2013
|
|
3O6O
 
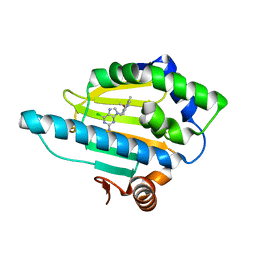 | | Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of an the inhibitor BIIB021 | | Descriptor: | 6-chloro-9-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-9H-purin-2-amine, Heat shock protein 83, PENTAETHYLENE GLYCOL | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, Weadge, J, Li, Y, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P, Fairlamb, A.H, Ferguson, M.A.J, Thompson, S, MacKenzie, C, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
2W2R
 
 | | Structure of the vesicular stomatitis virus matrix protein | | Descriptor: | MATRIX PROTEIN | | Authors: | Graham, S.C, Assenberg, R, Delmas, O, Verma, A, Gholami, A, Talbi, C, Owens, R.J, Stuart, D.I, Grimes, J.M, Bourhy, H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Rhabdovirus Matrix Protein Structures Reveal a Novel Mode of Self-Association.
Plos Pathog., 4, 2008
|
|
2VZ0
 
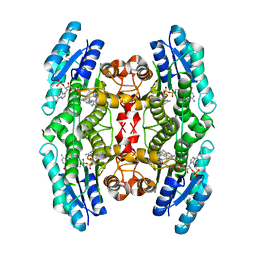 | | Pteridine Reductase 1 (PTR1) from Trypanosoma Brucei in complex with NADP and DDD00066641 | | Descriptor: | 6-(4-methylphenyl)quinazoline-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE | | Authors: | Robinson, D.A, Thompson, S, Sienkiewicz, N, Fairlamb, A.H. | | Deposit date: | 2008-07-29 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development and Validation of a Cytochrome C Coupled Assay for Pteridine Reductase 1 and Dihydrofolate Reductase.
Anal.Biochem., 396, 2010
|
|
2W2S
 
 | | Structure of the Lagos bat virus matrix protein | | Descriptor: | MATRIX PROTEIN | | Authors: | Graham, S.C, Assenberg, R, Delmas, O, Verma, A, Gholami, A, Talbi, C, Owens, R.J, Stuart, D.I, Grimes, J.M, Bourhy, H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rhabdovirus Matrix Protein Structures Reveal a Novel Mode of Self-Association.
Plos Pathog., 4, 2008
|
|
2WBA
 
 | | Properties of Trypanothione Reductase From T. brucei | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Jones, D, Ariza, A, Chow, W.H.A, Oza, S.L, Fairlamb, A.H. | | Deposit date: | 2009-02-24 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparative Structural, Kinetic and Inhibitor Studies of Trypanosoma Brucei Trypanothione Reductase with T. Cruzi.
Mol.Biochem.Parasitol., 169, 2010
|
|
3MX1
 
 | |
3MX4
 
 | | DNA binding and cleavage by the GIY-YIG endonuclease R.Eco29KI inactive variant E142Q | | Descriptor: | DNA (5'-D(P*CP*GP*GP*GP*AP*GP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*CP*GP*CP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*CP*GP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*CP*TP*CP*CP*CP*G)-3'), Eco29kIR | | Authors: | Mak, A.N.S, Lambert, A.R, Stoddard, B.L. | | Deposit date: | 2010-05-06 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Folding, DNA Recognition, and Function of GIY-YIG Endonucleases: Crystal Structures of R.Eco29kI.
Structure, 18, 2010
|
|
