1KV3
 
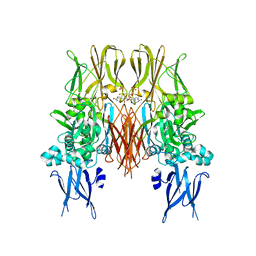 | | HUMAN TISSUE TRANSGLUTAMINASE IN GDP BOUND FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Protein-glutamine gamma-glutamyltransferase | | Authors: | Liu, S, Cerione, R.A, Clardy, J. | | Deposit date: | 2002-01-24 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8EGD
 
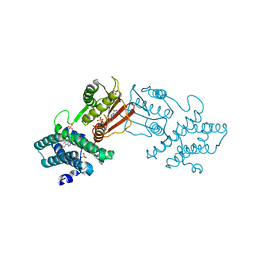 | | Branched chain ketoacid dehydrogenase kinase in complex with inhibitor | | Descriptor: | 5-(4-methoxyphenyl)-1H-tetrazole, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGQ
 
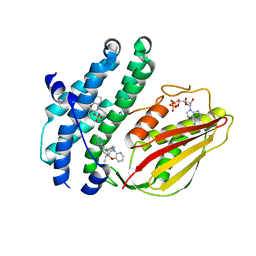 | | Branched chain ketoacid dehydrogenase kinase complexes | | Descriptor: | (2S)-2-ethyl-4-{[(2'M)-2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydro-2H-pyrido[3,2-b][1,4]oxazine, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGF
 
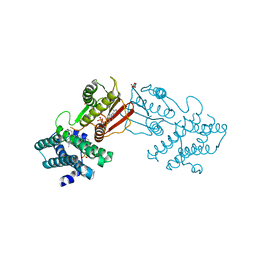 | | Branched chain ketoacid dehydrogenase kinase in complex with inhibitor | | Descriptor: | (5P)-5-(4'-methyl[1,1'-biphenyl]-2-yl)-1H-tetrazole, ADENOSINE-5'-DIPHOSPHATE, POTASSIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8G9V
 
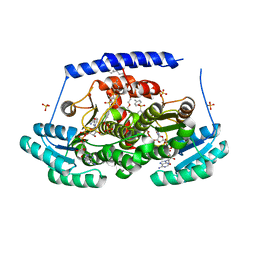 | | Crystal structures of 17-beta-hydroxysteroid dehydrogenase 13 | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 13, 4-{[2,5-dimethyl-3-(4-methylbenzene-1-sulfonyl)benzene-1-sulfonyl]amino}benzoic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Liu, S. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | Authors: | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | Deposit date: | 2015-04-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
1LL1
 
 | | HYDROXO BRIDGE MET FORM HEMOCYANIN FROM LIMULUS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, METCYANIN II | | Authors: | Liu, S, Magnus, K. | | Deposit date: | 1997-03-17 | | Release date: | 1997-08-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic Studies of Hydroxo, Nitrosyl, Azido and Fluro met Form Hemocyanin Subunit II from Limulus
To be Published
|
|
5J95
 
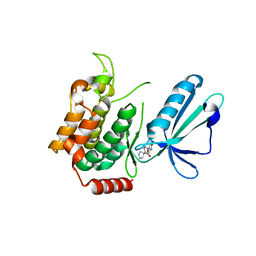 | | MAP4K4 in complex with inhibitor | | Descriptor: | 1-{4-[6-amino-5-(4-chlorophenyl)pyridin-3-yl]phenyl}cyclopentane-1-carboxylic acid, Mitogen-activated protein kinase kinase kinase kinase 4 | | Authors: | Liu, S. | | Deposit date: | 2016-04-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MAP4K4 in complex with inhibitor
To Be Published
|
|
3EB1
 
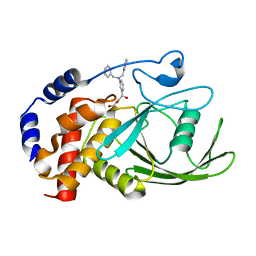 | | Crystal structure PTP1B complex with small molecule inhibitor LZP-25 | | Descriptor: | 4-[3-(dibenzylamino)phenyl]-2,4-dioxobutanoic acid, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Liu, S, Zheng, L.-F, Wu, L, Yu, X, Xue, T, Gunawan, A.M, Long, Y.-Q, Zhang, Z.-Y. | | Deposit date: | 2008-08-26 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting inactive enzyme conformation: aryl diketoacid derivatives as a new class of PTP1B inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
3TG2
 
 | | Crystal structure of the ISC domain of VibB in complex with isochorismate | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, TRIETHYLENE GLYCOL, Vibriobactin-specific isochorismatase | | Authors: | Liu, S, Zhang, C, Niu, B, Li, N, Liu, X, Liu, M, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structural insight into the ISC domain of VibB from Vibrio cholerae at atomic resolution: a snapshot just before the enzymatic reaction
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4TN6
 
 | | CK1d in complex with inhibitor | | Descriptor: | 4-{4-(4-fluorophenyl)-1-[1-(1,2-oxazol-3-ylmethyl)piperidin-4-yl]-1H-imidazol-5-yl}pyrimidin-2-amine, Casein kinase I isoform delta, SULFATE ION | | Authors: | Liu, S. | | Deposit date: | 2014-06-03 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | CK1D IN COMPLEX WITH inhibitor
To Be Published
|
|
3TB4
 
 | | Crystal structure of the ISC domain of VibB | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, S, Zhang, C, Niu, B, Li, N, Liu, M, Wei, T, Zhu, D, Xu, S, Gu, L. | | Deposit date: | 2011-08-05 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insight into the ISC domain of VibB from Vibrio cholerae at atomic resolution: a snapshot just before the enzymatic reaction
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8G93
 
 | | Crystal structures of 17-beta-hydroxysteroid dehydrogenase 13 | | Descriptor: | 3-fluoro-N-({(1r,4r)-4-[(2-fluorophenoxy)methyl]-1-hydroxycyclohexyl}methyl)-4-hydroxybenzamide, Hydroxysteroid 17-beta dehydrogenase 13, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, S. | | Deposit date: | 2023-02-21 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
8G84
 
 | | Crystal structures of HSD17B13 complexes | | Descriptor: | Hydroxysteroid 17-beta dehydrogenase 13, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, S. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.467 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
8G89
 
 | | HSD17B13 in complex with cofactor and inhibitor | | Descriptor: | 3-fluoro-N-({(1r,4r)-4-[(2-fluorophenoxy)methyl]-1-hydroxycyclohexyl}methyl)-4-hydroxybenzamide, Hydroxysteroid 17-beta dehydrogenase 13, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, S. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
1M1B
 
 | | Crystal Structure of Phosphoenolpyruvate Mutase Complexed with Sulfopyruvate | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE PHOSPHOMUTASE, SULFOPYRUVATE | | Authors: | Liu, S, Lu, Z, Jia, Y, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2002-06-18 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dissociative phosphoryl transfer in PEP mutase catalysis: structure of the enzyme/sulfopyruvate complex and kinetic properties of mutants.
Biochemistry, 41, 2002
|
|
4YHU
 
 | | Yeast Prp3 C-terminal fragment 296-469 | | Descriptor: | U4/U6 small nuclear ribonucleoprotein PRP3, YTTRIUM (III) ION | | Authors: | Liu, S, Wahl, M.C. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing.
Elife, 4, 2015
|
|
1BOA
 
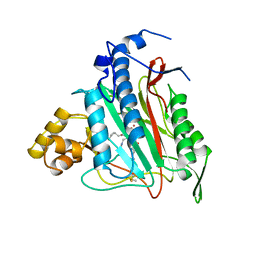 | | HUMAN METHIONINE AMINOPEPTIDASE 2 COMPLEXED WITH ANGIOGENESIS INHIBITOR FUMAGILLIN | | Descriptor: | COBALT (II) ION, FUMAGILLIN, METHIONINE AMINOPEPTIDASE | | Authors: | Liu, S, Widom, J, Kemp, C.W, Crews, C.M, Clardy, J.C. | | Deposit date: | 1998-08-01 | | Release date: | 1999-08-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
4B3Z
 
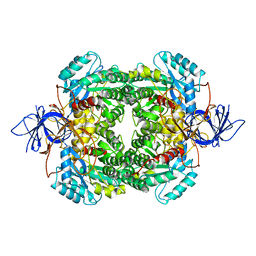 | | Structure of the human collapsin response mediator protein-1, a lung cancer suppressor | | Descriptor: | DIHYDROPYRIMIDINASE-RELATED PROTEIN 1 | | Authors: | Liu, S.H, Lin, Y.H, Huang, S.F, Niou, Y.K, Huang, L.L, Chen, Y.J. | | Deposit date: | 2012-07-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of Human Collapsin Response Mediator Protein 1: A Possible Role of its C-Terminal Tail.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8CUH
 
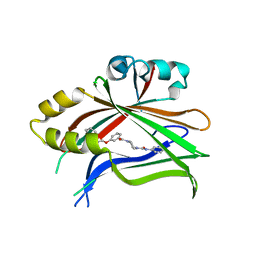 | | Crystal structure of human TEAD2 complexed with its inhibitor TM2. | | Descriptor: | 4-[3-(2-cyclohexylethoxy)benzoyl]-N-phenylpiperazine-1-carboxamide, Transcriptional enhancer factor TEF-4 | | Authors: | Liu, S, Luo, X. | | Deposit date: | 2022-05-17 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a new class of reversible TEA-domain transcription factor inhibitors with a novel binding mode.
Elife, 11, 2022
|
|
3JSW
 
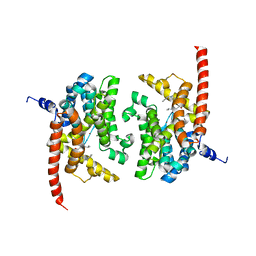 | | Human PDE9 in complex with selective inhibitor | | Descriptor: | 6-[(3S,4S)-1-benzyl-4-methylpyrrolidin-3-yl]-1-(1-methylethyl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2009-09-11 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a Brain Penetrant PDE9A Inhibitor Utilizing Prospective Design and Chemical Enablement as a Rapid Lead Optimization Strategy.
J.Med.Chem., 52, 2009
|
|
1D3G
 
 | | HUMAN DIHYDROOROTATE DEHYDROGENASE COMPLEXED WITH BREQUINAR ANALOG | | Descriptor: | 2-BIPHENYL-4-YL-6-FLUORO-3-METHYL-QUINOLINE-4-CARBOXYLIC ACID, ACETATE ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Liu, S, Neidhardt, E.A, Grossman, T.H, Ocain, T, Clardy, J. | | Deposit date: | 1999-09-29 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents.
Structure Fold.Des., 8, 2000
|
|
1OQF
 
 | | Crystal structure of the 2-methylisocitrate lyase | | Descriptor: | 2-methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Dunaway-Mariano, D, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-03-08 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 2-methylisocitrate lyase in complex with product and with isocitrate inhibitor provide insight into lyase substrate specificity, catalysis and evolution.
Biochemistry, 44, 2005
|
|
1D3H
 
 | | HUMAN DIHYDROOROTATE DEHYDROGENASE COMPLEXED WITH ANTIPROLIFERATIVE AGENT A771726 | | Descriptor: | (2Z)-2-cyano-3-hydroxy-N-[4-(trifluoromethyl)phenyl]but-2-enamide, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE, ... | | Authors: | Liu, S, Neidhardt, E.A, Grossman, T.H, Ocain, T, Clardy, J. | | Deposit date: | 1999-09-29 | | Release date: | 2000-08-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents.
Structure Fold.Des., 8, 2000
|
|
4M0V
 
 | | Crystal structure of E.coli SbcD with Mn2+ | | Descriptor: | Exonuclease subunit SbcD, GLYCEROL, MANGANESE (II) ION | | Authors: | Liu, S, Tian, L.F, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-08-02 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
