3LKZ
 
 | |
3V0J
 
 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S), Deletion of 401-405 | | Descriptor: | PHOSPHATE ION, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V0E
 
 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 256-576(C363S) | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V0H
 
 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S), complexed with D-MYO-inositol-1,4,5-triphosphate | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V0F
 
 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S), form II | | Descriptor: | PHOSPHATE ION, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7T42
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 2c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[2-(2-methylpropanoyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(2-acetyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T44
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 4c | | Descriptor: | (1R,2S)-2-[(N-{[(2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[2-(methanesulfonyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T43
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | Descriptor: | (1R,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
1T2L
 
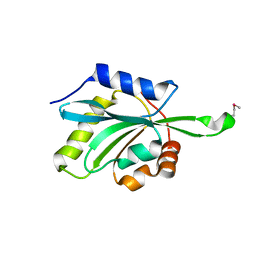 | | Three Crystal Structures of Human Coactosin-like Protein | | Descriptor: | Coactosin-like protein | | Authors: | Liu, L, Wei, Z, Chen, Z, Wang, Y, Gong, W. | | Deposit date: | 2004-04-22 | | Release date: | 2004-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Coactosin-like Protein
J.Mol.Biol., 344, 2004
|
|
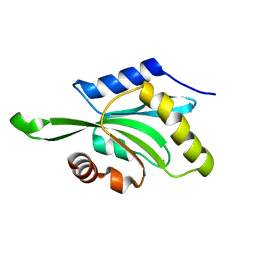
1T3X
 
 | |
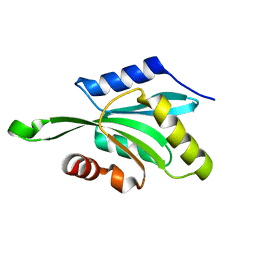
1T3Y
 
 | |
6C4X
 
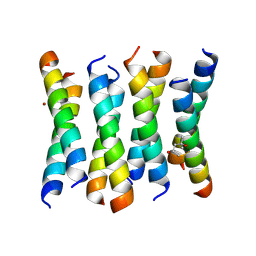 | | Cross-alpha Amyloid-like Structure alphaAmmem | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ZINC ION, cross-alpha amyloid-like membrane peptide alpha-AmMEM | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C50
 
 | | Cross-alpha Amyloid-like Structure alphaAmS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmS, FORMIC ACID | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C51
 
 | | Cross-alpha Amyloid-like Structure alphaAmL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmL, PHOSPHATE ION | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
7TQ8
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 14d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, Orf1a protein, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
3V0I
 
 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 256-576, E411F | | Descriptor: | SULFATE ION, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7YP2
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement) | | Descriptor: | 6H2 heavy chain, 6H2 light chain, Envelope glycoprotein H | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
8IJC
 
 | | NMR solution structure of the 1:1 complex of a platinum(II) ligand L1-transpt covalently bound to a G-quadruplex MYT1L | | Descriptor: | G-quadruplex DNA MYT1L, Pt(NH3)2(2-(pyridin-4-ylmethyl)benzo-[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) | | Authors: | Liu, L.-Y, Mao, Z.-W. | | Deposit date: | 2023-02-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Organic-Platinum Hybrids for Covalent Binding of G-Quadruplexes: Structural Basis and Application to Cancer Immunotherapy.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6JW0
 
 | | Universal RVD R* accommodates cytosine via water-mediated interactions | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*CP*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
2B3R
 
 | | Crystal structure of the C2 domain of class II phosphatidylinositide 3-kinase C2 | | Descriptor: | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide, SULFATE ION | | Authors: | Liu, L, Song, X, He, D, Komma, C, Kita, A, Verbasius, J.V, Bellamy, H, Miki, K, Czech, M.P, Zhou, G.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the C2 domain of class II phosphatidylinositide 3-kinase C2alpha.
J.Biol.Chem., 281, 2006
|
|
7YP1
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement) | | Descriptor: | 10E4 heavy chain, 10E4 light chain, EBV gH, ... | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YOY
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement) | | Descriptor: | 3E8 heavy chain, 3E8 light chain, 5E3 heavy chain, ... | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
6C52
 
 | | Cross-alpha Amyloid-like Structure alphaTet | | Descriptor: | Cross-alpha Amyloid-like Structure alphaTet, GLYCEROL | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
7SGL
 
 | | DNA-PK complex of DNA end processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, Hairpin_1, ... | | Authors: | Liu, L, Li, J, Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2021-10-06 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
3GUP
 
 | |
