7WXH
 
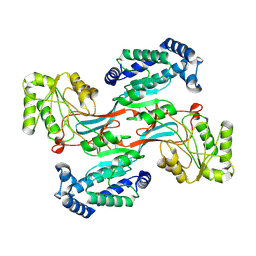 | | GPR domain open form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WXG
 
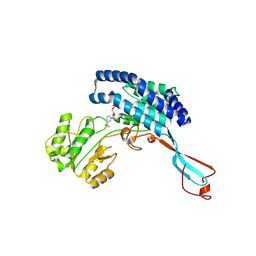 | | GPR domain closed form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WXF
 
 | |
7WX3
 
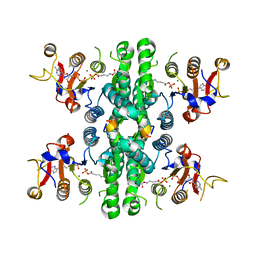 | | GK domain of Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase, GAMMA-GLUTAMYL PHOSPHATE, ... | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7CJB
 
 | |
8XHR
 
 | |
7V59
 
 | | Cryo-EM structure of spyCas9-sgRNA-DNA dimer | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (49-MER), RNA (115-MER) | | Authors: | Liu, J, Deng, P. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.26 Å) | | Cite: | Nonspecific interactions between SpCas9 and dsDNA sites located downstream of the PAM mediate facilitated diffusion to accelerate target search.
Chem Sci, 12, 2021
|
|
6IR8
 
 | | Rice WRKY/DNA complex | | Descriptor: | DNA (5'-D(P*GP*AP*TP*AP*TP*TP*TP*GP*AP*CP*CP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*GP*GP*TP*CP*AP*AP*AP*TP*AP*TP*C)-3'), OsWRKY45, ... | | Authors: | Liu, J, Cheng, X, Wang, D. | | Deposit date: | 2018-11-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of dimerization and dual W-box DNA recognition by rice WRKY domain.
Nucleic Acids Res., 47, 2019
|
|
4USG
 
 | | Crystal structure of PC4 W89Y mutant complex with DNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP *TP*TP*TP*TP*TP*G)-3', ACTIVATED RNA POLYMERASE II TRANSCRIPTIONAL COACTIVATOR P15 | | Authors: | Zhao, Y, Liu, J. | | Deposit date: | 2014-07-08 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4.
Sci.Rep., 5, 2015
|
|
1QA9
 
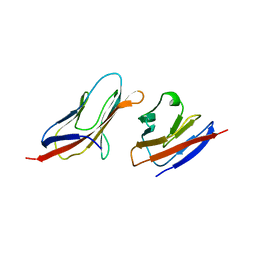 | | Structure of a Heterophilic Adhesion Complex Between the Human CD2 and CD58(LFA-3) Counter-Receptors | | Descriptor: | HUMAN CD2 PROTEIN, HUMAN CD58 PROTEIN | | Authors: | Wang, J.-H, Smolyar, A, Tan, K, Liu, J.-H, Kim, M, Sun, Z.J, Wagner, G, Reinherz, E.L. | | Deposit date: | 1999-04-13 | | Release date: | 1999-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors.
Cell(Cambridge,Mass.), 97, 1999
|
|
1H0J
 
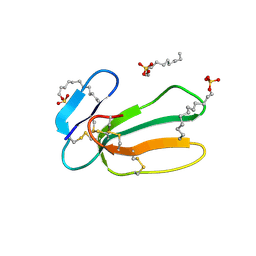 | | Structural Basis of the Membrane-induced Cardiotoxin A3 Oligomerization | | Descriptor: | CARDIOTOXIN-3, DODECYL SULFATE | | Authors: | Forouhar, F, Huang, W.-N, Liu, J.-H, Chien, K.-Y, Wu, W.-G, Hsiao, C.-D. | | Deposit date: | 2002-06-20 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Membrane-Induced Cardiotoxin A3 Oligomerization
J.Biol.Chem., 278, 2003
|
|
3C86
 
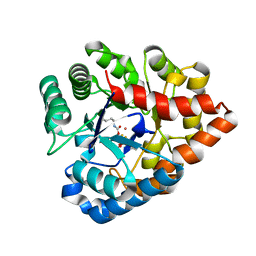 | | OpdA from agrobacterium radiobacter with bound product diethyl thiophosphate from crystal soaking with tetraethyl dithiopyrophosphate- 1.8 A | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, FE (II) ION, ... | | Authors: | Ollis, D.L, Jackson, C.J, Foo, J.L, Kim, H.K, Carr, P.D, Liu, J.W, Salem, G. | | Deposit date: | 2008-02-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase
J.Mol.Biol., 375, 2008
|
|
3C0B
 
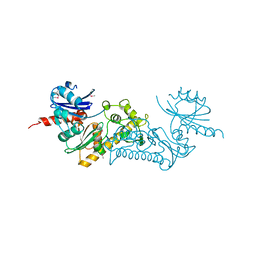 | | Crystal structure of the conserved archaeal protein Q6M145. Northeast Structural Genomics Consortium target MrR63 | | Descriptor: | CALCIUM ION, Conserved archaeal protein Q6M145 | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-19 | | Release date: | 2008-02-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of the conserved archaeal protein Q6M145.
To be Published
|
|
3C37
 
 | | X-ray structure of the putative Zn-dependent peptidase Q74D82 at the resolution 1.7 A. Northeast Structural Genomics Consortium target GsR143A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidase, M48 family, ... | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Vorobiev, S.M, Forouhar, F, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-27 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the putative Zn-dependent peptidase Q74D82 at the resolution 1.7 A.
To be Published
|
|
3K2Q
 
 | | Crystal structure of Pyrophosphate-dependent phosphofructokinase from Marinobacter aquaeolei, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET MqR88 | | Descriptor: | Pyrophosphate-dependent phosphofructokinase, SODIUM ION | | Authors: | Seetharaman, J, Lew, S, Wang, D, Neely, H, Janjua, K, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Pyrophosphate-dependent phosphofructokinase from Marinobacter aquaeolei, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET MqR88
To be Published
|
|
3K2T
 
 | | Crystal structure of Lmo2511 protein from Listeria monocytogenes, northeast structural genomics consortium target LkR84A | | Descriptor: | Lmo2511 protein | | Authors: | Seetharaman, J, Su, M, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Lmo2511 protein from Listeria monocytogenes, northeast structural genomics consortium target LkR84A
To be Published
|
|
3JC7
 
 | | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | | Descriptor: | Cell division control protein 45, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Li, H, Bai, L, Yuan, Z, Sun, J, Georgescu, R.E, Liu, J, O'Donnell, M.E. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3J8G
 
 | | Electron cryo-microscopy structure of EngA bound with the 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Zhang, X, Yan, K, Zhang, Y, Li, N, Ma, C, Li, Z, Zhang, Y, Feng, B, Liu, J, Sun, Y, Xu, Y, Lei, J, Gao, N. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly
Nucleic Acids Res., 2014
|
|
2A5I
 
 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in the space group C2 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 1,2-ETHANEDIOL, 3C-like peptidase, ... | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2A5K
 
 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in space group P212121 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 3C-like peptidase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
1ZV8
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | ACETATE ION, CACODYLATE ION, E2 glycoprotein, ... | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
3JCB
 
 | | Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5 | | Descriptor: | Antibody 36D5 heavy chain, Antibody 36D5 light chain, Envelope glycoprotein gp120, ... | | Authors: | Hu, G, Liu, J, Roux, K, Taylor, K.A. | | Deposit date: | 2015-11-25 | | Release date: | 2017-05-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structure of Simian Immunodeficiency Virus Envelope Spikes Bound with CD4 and Monoclonal Antibody 36D5.
J. Virol., 91, 2017
|
|
1ZVB
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | E2 glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
6VRV
 
 | | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model | | Descriptor: | 4-chloro-1-{(1S)-1-[(3S)-3-fluoropyrrolidin-3-yl]ethyl}-3-methyl-1H-pyrrolo[2,3-b]pyridine-6-carboxamide, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Barberis, C.E, Batchelor, J.D, Liu, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
1ZV7
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | CHLORIDE ION, spike glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
