4FA7
 
 | |
7XIC
 
 | | S-ECD (Omicron) in complex with STS165 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional analysis of an inter-Spike bivalent neutralizing antibody against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7XID
 
 | | S-ECD (Omicron) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional analysis of an inter-Spike bivalent neutralizing antibody against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
4FAA
 
 | |
6QPW
 
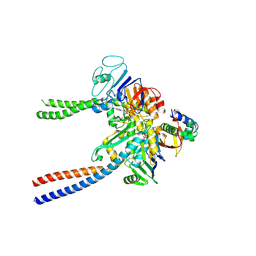 | | Structural basis of cohesin ring opening | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sister chromatid cohesion protein 1, ... | | Authors: | Panne, D, Muir, K.W, Li, Y, Weis, F. | | Deposit date: | 2019-02-15 | | Release date: | 2020-02-05 | | Last modified: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of the cohesin ATPase elucidates the mechanism of SMC-kleisin ring opening.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8BS6
 
 | |
6ZD4
 
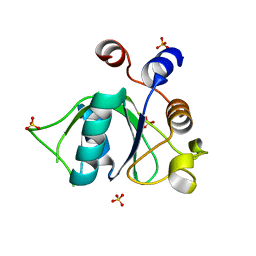 | | Crystal structure of YTHDC1 S378A mutant | | Descriptor: | SULFATE ION, YTH domain containing 1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2020-06-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZD3
 
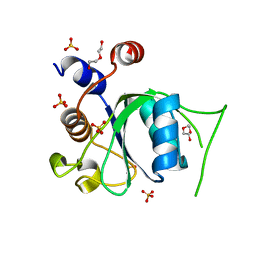 | | Crystal structure of YTHDC1 M438A mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, YTH domain containing 1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2020-06-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
2P91
 
 | | Crystal structure of Enoyl-[acyl-carrier-protein] reductase (NADH) from Aquifex aeolicus VF5 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Chen, L, Li, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Enoyl-[acyl-carrier-protein] reductase (NADH) from Aquifex aeolicus VF5
To be Published
|
|
6ZD5
 
 | |
6ZDA
 
 | |
6ZD8
 
 | | Crystal structure of YTHDC1 T379V mutant | | Descriptor: | SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
7SSE
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with CYCA-117-70 | | Descriptor: | DDB1- and CUL4-associated factor 1, N-[(3R)-1-(3-fluorophenyl)piperidin-3-yl]-6-(morpholin-4-yl)pyrimidin-4-amine | | Authors: | Kimani, S, Owen, J, Li, A, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Shahani, V.M, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of a Novel DCAF1 Ligand Using a Drug-Target Interaction Prediction Model: Generalizing Machine Learning to New Drug Targets.
J.Chem.Inf.Model., 63, 2023
|
|
5CP7
 
 | | Crystal Structure of an Antigen-Binding Fragment of Monoclonal Antibody against Sulfonamides | | Descriptor: | Heavy Chain of Antigen-Binding Fragment of Monoclonal Antibody of 4C7, Light Chain of Antigen-Binding Fragment of Monoclonal Antibody of 4C7 | | Authors: | Wang, Z, Shen, J, Li, C, Li, Y, Wen, K, Yu, X, Zhang, X. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Class-specific Monoclonal Antibodies and Dihydropteroate Synthase in Bioassays used for the Detection of Sulfonamides: Structural Insights into Recognition Diversity.
Anal. Chem., 91, 2019
|
|
1KAE
 
 | | L-HISTIDINOL DEHYDROGENASE (HISD) STRUCTURE COMPLEXED WITH L-HISTIDINOL (SUBSTRATE), ZINC AND NAD (COFACTOR) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Histidinol dehydrogenase, ... | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J.D, Cygler, M. | | Deposit date: | 2001-11-01 | | Release date: | 2002-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K75
 
 | | The L-histidinol dehydrogenase (hisD) structure implicates domain swapping and gene duplication. | | Descriptor: | GLYCEROL, L-histidinol dehydrogenase, SULFATE ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-10-18 | | Release date: | 2002-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KAH
 
 | | L-HISTIDINOL DEHYDROGENASE (HISD) STRUCTURE COMPLEXED WITH L-HISTIDINE (PRODUCT), ZN AND NAD (COFACTOR) | | Descriptor: | HISTIDINE, Histidinol dehydrogenase, ZINC ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J.D, Cygler, M. | | Deposit date: | 2001-11-02 | | Release date: | 2002-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5B73
 
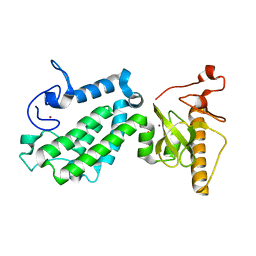 | | Crystal structure of human ZMYND8 PHD-Bromo-PWWP domain | | Descriptor: | Protein kinase C-binding protein 1, ZINC ION | | Authors: | Li, H, Li, Y, Zheng, X. | | Deposit date: | 2016-06-03 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ZMYND8 Reads the Dual Histone Mark H3K4me1-H3K14ac to Antagonize the Expression of Metastasis-Linked Genes
Mol.Cell, 63, 2016
|
|
1K6M
 
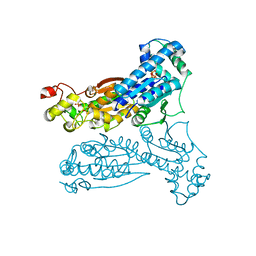 | | Crystal Structure of Human Liver 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase | | Descriptor: | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2-phosphatase, PHOSPHATE ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Lee, Y.H, Li, Y, Uyeda, K, Hasemann, C.A. | | Deposit date: | 2001-10-16 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tissue-specific structure/function differentiation of the liver isoform of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase.
J.Biol.Chem., 278, 2003
|
|
7RUH
 
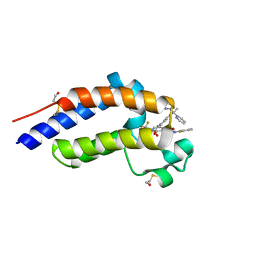 | | Bromodomain-containing protein 4 (BRD4) bromodomain 2 (BD2) complexed with XR844 | | Descriptor: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(5-{[(2-fluorophenyl)carbamoyl]amino}-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-1H-indol-4-yl}-2,2,2-trifluoroethane-1-sulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bromodomain-containing protein 4 (BRD4) bromodomain 2 (BD2) complexed with XR844
To Be Published
|
|
7RUI
 
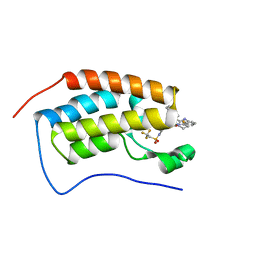 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with XR844 | | Descriptor: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(5-{[(2-fluorophenyl)carbamoyl]amino}-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-1H-indol-4-yl}-2,2,2-trifluoroethane-1-sulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with XR844
To Be Published
|
|
5XI4
 
 | | BRD4 bound with compound Bdi4 | | Descriptor: | (3~{S})-4-cyclopropyl-1,3-dimethyl-6-[[(1~{S})-1-(4-methylphenyl)ethyl]amino]-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D, Li, Y. | | Deposit date: | 2017-04-25 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | BRD4 bound with compound Bdi4
To Be Published
|
|
8Q72
 
 | | E. coli plasmid-borne JetABCD(E248A) core in a cleavage-competent state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circular plasmid DNA (1840-MER), JetA, ... | | Authors: | Roisne-Hamelin, F, Li, Y, Gruber, S. | | Deposit date: | 2023-08-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural basis for plasmid restriction by SMC JET nuclease.
Mol.Cell, 84, 2024
|
|
5XI3
 
 | | BRD4 bound with compound Bdi3 | | Descriptor: | (3~{R})-4-cyclopropyl-1,3-dimethyl-6-[[(1~{R})-1-phenylethyl]amino]-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D, Li, Y. | | Deposit date: | 2017-04-25 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.674 Å) | | Cite: | BRD4 bound with compound Bdi3
To Be Published
|
|
8Q2Q
 
 | | Crystal structure of YTHDC1 in complex with Compound 2b (YL_32) | | Descriptor: | 2-chloranyl-~{N},9-dimethyl-purin-6-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2023-08-03 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Based Design of a Potent and Selective YTHDC1 Ligand.
J.Med.Chem., 67, 2024
|
|
