7JKY
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with YF3-126 | | Descriptor: | Bromodomain-containing protein 4, N-(1-[1,1-di(pyridin-2-yl)ethyl]-6-{1-methyl-6-oxo-5-[(piperidin-4-yl)amino]-1,6-dihydropyridin-3-yl}-1H-indol-4-yl)ethanesulfonamide, SODIUM ION | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
7JKZ
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 2 (BD2) complexed with YF3-126 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
7JKW
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with ZN1-99 | | Descriptor: | Bromodomain-containing protein 4, N-(6-{5-[(azetidin-3-yl)amino]-1-methyl-6-oxo-1,6-dihydropyridin-3-yl}-1-[1,1-di(pyridin-2-yl)ethyl]-1H-indol-4-yl)ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
7JKX
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with YF3-6 | | Descriptor: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-[1-methyl-5-(methylamino)-6-oxo-1,6-dihydropyridin-3-yl]-1H-indol-4-yl}ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with YF3-6
To Be Published
|
|
6A3E
 
 | | MVM NES mutant Nm15 in complex with CRM1-Ran-RanBP1 | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sun, Q, Li, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cancer Therapy with Nanoparticle-Medicated Intracellular Expression of Peptide CRM1-Inhibitor.
Int J Nanomedicine, 16, 2021
|
|
7L97
 
 | | Crystal structure of STAMBPL1 in complex with an engineered binder | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, SULFATE ION, ... | | Authors: | Guo, Y, Dong, A, Hou, F, Li, Y, Zhang, W, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of ubiquitin variant inhibitors for the JAMM-family deubiquitinases STAMBP and STAMBPL1.
J.Biol.Chem., 297, 2021
|
|
6A3C
 
 | | MVM NES mutant Nm12 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Li, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cancer Therapy with Nanoparticle-Medicated Intracellular Expression of Peptide CRM1-Inhibitor.
Int J Nanomedicine, 16, 2021
|
|
1A4I
 
 | | HUMAN TETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE | | Descriptor: | METHYLENETETRAHYDROFOLATE DEHYDROGENASE / METHENYLTETRAHYDROFOLATE CYCLOHYDROLASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Allaire, M, Li, Y, Mackenzie, R.E, Cygler, M. | | Deposit date: | 1998-01-30 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 3-D structure of a folate-dependent dehydrogenase/cyclohydrolase bifunctional enzyme at 1.5 A resolution.
Structure, 6, 1998
|
|
4ER3
 
 | | Crystal Structure of Human DOT1L in complex with inhibitor EPZ004777 | | Descriptor: | 1,2-ETHANEDIOL, 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-lysine N-methyltransferase, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
6A38
 
 | | MVM NS2 NES in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, Exportin-1, GLYCEROL, ... | | Authors: | Sun, Q, Li, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Cancer Therapy with Nanoparticle-Medicated Intracellular Expression of Peptide CRM1-Inhibitor.
Int J Nanomedicine, 16, 2021
|
|
4ER6
 
 | | Crystal structure of human DOT1L in complex with inhibitor SGC0946 | | Descriptor: | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, BROMIDE ION, Histone-lysine N-methyltransferase, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
6A30
 
 | | Crystal Structure of Munc13-1 MUN Domain and Synaptobrevin-2 Juxtamembrane Linker Region | | Descriptor: | Protein unc-13 homolog A, Synaptobrevin-2 juxtamembrane linker peptide | | Authors: | Wang, S, Li, Y, Gong, J.H, Ye, S, Yang, X.F, Zhang, R.G, Ma, C. | | Deposit date: | 2018-06-14 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Munc18 and Munc13 serve as a functional template to orchestrate neuronal SNARE complex assembly.
Nat Commun, 10, 2019
|
|
6A3B
 
 | | MVM NES mutant Nm13 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, Exportin-1, GLYCEROL, ... | | Authors: | Sun, Q, Li, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Cancer Therapy with Nanoparticle-Medicated Intracellular Expression of Peptide CRM1-Inhibitor.
Int J Nanomedicine, 16, 2021
|
|
5YUF
 
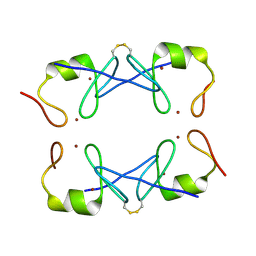 | | Crystal Structure of PML RING tetramer | | Descriptor: | Protein PML, ZINC ION | | Authors: | Wang, P, Benhend, S, Wu, H, Breitenbach, V, Zhen, T, Jollivet, F, Peres, L, Li, Y, Chen, S, Chen, Z, de THE, H, Meng, G. | | Deposit date: | 2017-11-22 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RING tetramerization is required for nuclear body biogenesis and PML sumoylation.
Nat Commun, 9, 2018
|
|
7XYT
 
 | |
7YFG
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (major class in asymmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFH
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine, glutamate and (R)-PYD-106 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7W3Q
 
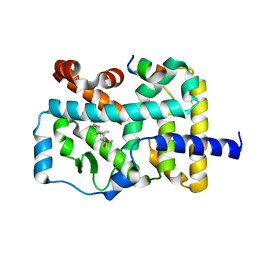 | | Crystal structure of RORgamma in complex with natural inverse agonist | | Descriptor: | (3S,5R,6S,8R,9R,10R,12R,13R,14R,17S)-4,4,8,10,14-pentamethyl-17-[(2R)-6-methyl-2-oxidanyl-hept-5-en-2-yl]-2,3,5,6,7,9,11,12,13,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthrene-3,6,12-triol, Nuclear receptor ROR-gamma, Peptide from Nuclear receptor coactivator 2 | | Authors: | Tian, S.Y, Li, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of RORgamma in complex with natural inverse agonist
To Be Published
|
|
7W3P
 
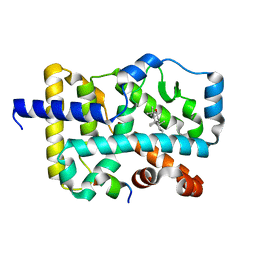 | | Crystal structure of RORgamma in complex with natural inverse agonist | | Descriptor: | (3S,5R,8R,9R,10R,12R,13R,14R,17S)-4,4,8,10,14-pentamethyl-17-[(2R)-2,6,6-trimethyloxan-2-yl]-2,3,5,6,7,9,11,12,13,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthrene-3,12-diol, Nuclear receptor ROR-gamma, Peptide from Nuclear receptor coactivator 2 | | Authors: | Tian, S.Y, Li, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of RORgamma in complex with natural inverse agonist
To Be Published
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2PYE
 
 | | Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC RevealNative Diagonal Binding Geometry TCR Clone C5C1 Complexed with MHC | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Beta-2-microglobulin, Cancer/testis antigen 1B, ... | | Authors: | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | Deposit date: | 2007-05-16 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
6I6N
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
6ISA
 
 | | mCD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6ISB
 
 | | crystal structure of human CD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7ETW
 
 | | Cryo-EM structure of Scap/Insig complex in the present of digitonin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Digitonin, Insulin-induced gene 2 protein, ... | | Authors: | Yan, R, Cao, P, Song, W, Li, Y, Wang, T, Qian, H, Yan, C, Yan, N. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-23 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for sterol sensing by Scap and Insig
Cell Rep, 35, 2021
|
|
