5UO8
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 3-[(2-aminoquinolin-7-yl)methoxy]-5-((methylamino)methyl)benzonitrile | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(2-aminoquinolin-7-yl)methoxy]-5-[(methylamino)methyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
5CVE
 
 | |
2IS5
 
 | |
2IKB
 
 | |
6VSI
 
 | | Crystal structure of FKBP12 of Candida auris | | Descriptor: | Peptidylprolyl isomerase, SULFATE ION | | Authors: | Li, Z, Li, H, Hernandez, G, LeMaster, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure and transient dimerization for the FKBP12 protein from the pathogenic fungus Candida auris.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
4ILM
 
 | | CRISPR RNA Processing endoribonuclease | | Descriptor: | CRISPR-associated endoribonuclease Cas6 2, RNA (5'-R(*GP*CP*UP*AP*AP*UP*CP*UP*AP*CP*UP*AP*UP*AP*GP*A)-3') | | Authors: | Shao, Y, Li, H. | | Deposit date: | 2012-12-31 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.068 Å) | | Cite: | Recognition and cleavage of a nonstructured CRISPR RNA by its processing endoribonuclease Cas6.
Structure, 21, 2013
|
|
6IG9
 
 | |
6W6G
 
 | |
6W6H
 
 | |
6W6E
 
 | |
6W6J
 
 | |
6W6I
 
 | |
4W5A
 
 | | Complex structure of ATRX ADD bound to H3K9me3S10ph peptide | | Descriptor: | Peptide from Histone H3.3, Transcriptional regulator ATRX, ZINC ION | | Authors: | Zhao, D, Xiang, B, Li, H. | | Deposit date: | 2014-08-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3EFA
 
 | | Crystal structure of putative N-acetyltransferase from Lactobacillus plantarum | | Descriptor: | FORMIC ACID, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chang, C, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Crystal structure of putative N-acetyltransferase from Lactobacillus plantarum
To be Published
|
|
2I6D
 
 | | The structure of a putative RNA methyltransferase of the TrmH family from Porphyromonas gingivalis. | | Descriptor: | ACETIC ACID, RNA methyltransferase, TrmH family | | Authors: | Cuff, M.E, Mussar, K.E, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-28 | | Release date: | 2006-10-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of a putative RNA methyltransferase of the TrmH family from Porphyromonas gingivalis.
TO BE PUBLISHED
|
|
2I9D
 
 | |
6POW
 
 | | Structure of human endotheial nitric oxide synthase heme domain in complex with 7-(5-(Aminomethyl)pyridin-3-yl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[5-(aminomethyl)pyridin-3-yl]-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
8IHS
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHR
 
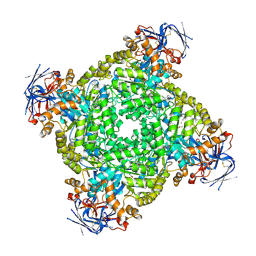 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | Descriptor: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHQ
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 | | Descriptor: | Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
6POZ
 
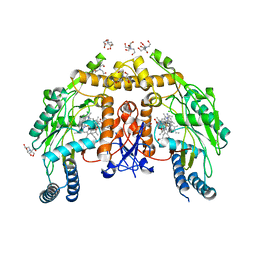 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-isopropoxyphenyl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-{3-(aminomethyl)-4-[(propan-2-yl)oxy]phenyl}-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
8ITM
 
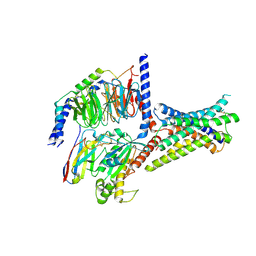 | | Cryo-EM structure of GIPR splice variant 2 (SV2) in complex with Gs protein | | Descriptor: | Gastric inhibitory polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Hang, K.N, Zhou, Q.T, Shao, L.J, Li, H, Li, W.Z, Lin, S, Dai, A.T, Cai, X.Q, Liu, Y.Y, Xu, Y.N, Feng, W.B, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-22 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Molecular basis of signal transduction mediated by the human GIPR splice variants.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ITL
 
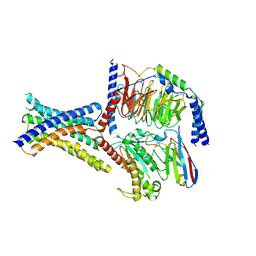 | | Cryo-EM structure of GIPR splice variant 1 (SV1) in complex with Gs protein | | Descriptor: | Gastric inhibitory polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Hang, K.N, Zhou, Q.T, Shao, L.J, Li, H, Li, W.Z, Lin, S, Dai, A.T, Cai, X.Q, Liu, Y.Y, Xu, Y.N, Feng, W.B, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-22 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular basis of signal transduction mediated by the human GIPR splice variants.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6POX
 
 | | Structure of human endothelialnitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-ethoxyphenyl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-ethoxyphenyl]-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6POY
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-propoxyphenyl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-propoxyphenyl]-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
