3GU5
 
 | |
4CPH
 
 | | trans-divalent streptavidin with love-hate ligand 4 | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N'-{2,6-bis[4-(morpholine-4- sulfonyl)phenyl]phenyl}pentanehydrazide, STREPTAVIDIN | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
4CPI
 
 | | streptavidin A86D mutant with love-hate ligand 4 | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N'-{2,6-bis[4-(morpholine-4- sulfonyl)phenyl]phenyl}pentanehydrazide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
4CPF
 
 | | Wild-type streptavidin in complex with love-hate ligand 3 (LH3) | | Descriptor: | STREPTAVIDIN, methyl 4-(2-{5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H- thieno[3,4-d]imidazolidin-4-yl]pentanehydrazido}-3- [4-(methoxycarbonyl)phenyl]phenyl)benzoate | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
3GU4
 
 | |
4CPE
 
 | | Wild-type streptavidin in complex with love-hate ligand 1 (LH1) | | Descriptor: | (3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N-{2-[(2,6- diphenylphenyl)formamido]ethyl}pentanamide, STREPTAVIDIN | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
3HDH
 
 | | PIG HEART SHORT CHAIN L-3-HYDROXYACYL COA DEHYDROGENASE REVISITED: SEQUENCE ANALYSIS AND CRYSTAL STRUCTURE DETERMINATION | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (L-3-HYDROXYACYL COA DEHYDROGENASE) | | Authors: | Barycki, J.J, O'Brien, L.K, Birktoft, J.J, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 1999-04-13 | | Release date: | 1999-10-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pig heart short chain L-3-hydroxyacyl-CoA dehydrogenase revisited: sequence analysis and crystal structure determination.
Protein Sci., 8, 1999
|
|
3F5G
 
 | | Crystal structure of death associated protein kinase in complex with ADP and Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Death-associated protein kinase 1, MAGNESIUM ION | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-11-03 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3K5B
 
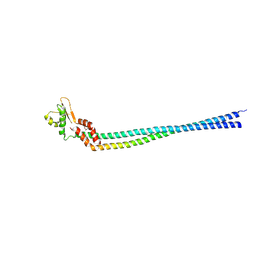 | | Crystal structure of the peripheral stalk of Thermus thermophilus H+-ATPase/synthase | | Descriptor: | V-type ATP synthase subunit E, V-type ATP synthase, subunit (VAPC-THERM) | | Authors: | Lee, L.K, Stewart, A.G, Donohoe, M, Bernal, R.A, Stock, D. | | Deposit date: | 2009-10-07 | | Release date: | 2010-02-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of the peripheral stalk of Thermus thermophilus H(+)-ATPase/synthase.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1F0Y
 
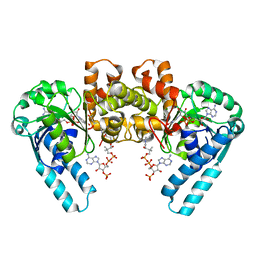 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH ACETOACETYL-COA AND NAD+ | | Descriptor: | ACETOACETYL-COENZYME A, L-3-HYDROXYACYL-COA DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2000-05-17 | | Release date: | 2000-09-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
1F3T
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA BRUCEI ORNITHINE DECARBOXYLASE (ODC) COMPLEXED WITH PUTRESCINE, ODC'S REACTION PRODUCT. | | Descriptor: | 1,4-DIAMINOBUTANE, ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jackson, L.K, Brooks, H.B, Osterman, A.L, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Altering the reaction specificity of eukaryotic ornithine decarboxylase.
Biochemistry, 39, 2000
|
|
1F17
 
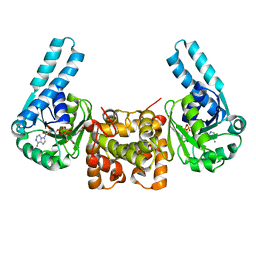 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-3-HYDROXYACYL-COA DEHYDROGENASE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2000-05-18 | | Release date: | 2000-09-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
1F12
 
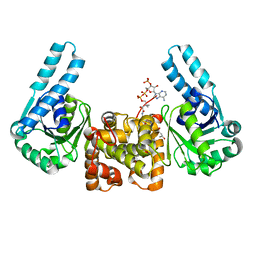 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH 3-HYDROXYBUTYRYL-COA | | Descriptor: | 3-HYDROXYBUTANOYL-COENZYME A, L-3-HYDROXYACYL-COA DEHYDROGENASE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2000-05-18 | | Release date: | 2000-09-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
1F0Z
 
 | | SOLUTION STRUCTURE OF THIS, THE SULFUR CARRIER PROTEIN IN E.COLI THIAMIN BIOSYNTHESIS | | Descriptor: | THIS PROTEIN | | Authors: | Wang, C, Xi, J, Begley, T.P, Nicholson, L.K. | | Deposit date: | 2000-05-17 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ThiS and implications for the evolutionary roots of ubiquitin.
Nat.Struct.Biol., 8, 2001
|
|
1LKS
 
 | | HEN EGG WHITE LYSOZYME NITRATE | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Steinrauf, L.K. | | Deposit date: | 1997-10-09 | | Release date: | 1998-04-29 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures of monoclinic lysozyme iodide at 1.6 A and of triclinic lysozyme nitrate at 1.1 A.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1LKR
 
 | | MONOCLINIC HEN EGG WHITE LYSOZYME IODIDE | | Descriptor: | IODIDE ION, LYSOZYME | | Authors: | Steinrauf, L.K. | | Deposit date: | 1998-01-20 | | Release date: | 1998-04-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of monoclinic lysozyme iodide at 1.6 A and of triclinic lysozyme nitrate at 1.1 A.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1BSO
 
 | | 12-BROMODODECANOIC ACID BINDS INSIDE THE CALYX OF BOVINE BETA-LACTOGLOBULIN | | Descriptor: | 12-BROMODODECANOIC ACID, PROTEIN (BOVINE BETA-LACTOGLOBULIN A) | | Authors: | Qin, B.Y, Creamer, L.K, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-29 | | Release date: | 1999-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | 12-Bromododecanoic acid binds inside the calyx of bovine beta-lactoglobulin.
FEBS Lett., 438, 1998
|
|
1BSY
 
 | | STRUCTURAL BASIS OF THE TANFORD TRANSITION OF BOVINE BETA-LACTOGLOBULIN FROM CRYSTAL STRUCTURES AT THREE PH VALUES; PH 7.1 | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Qin, B.Y, Bewley, M.C, Creamer, L.K, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-31 | | Release date: | 1999-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis of the Tanford transition of bovine beta-lactoglobulin.
Biochemistry, 37, 1998
|
|
1LY1
 
 | |
1BSQ
 
 | | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF POINT MUTATIONS OF VARIANTS A AND B OF BOVINE BETA-LACTOGLOBULIN | | Descriptor: | PROTEIN (BETA-LACTOGLOBULIN) | | Authors: | Qin, B.Y, Creamer, L.K, Bewley, M.C, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-29 | | Release date: | 1998-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Functional implications of structural differences between variants A and B of bovine beta-lactoglobulin.
Protein Sci., 8, 1999
|
|
1M6O
 
 | | Crystal Structure of HLA B*4402 in complex with HLA DPA*0201 peptide | | Descriptor: | Beta-2-microglobulin, HLA DPA*0201 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N.A, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | Deposit date: | 2002-07-17 | | Release date: | 2003-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
1D4K
 
 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | HIV-1 PROTEASE, N-13-[(10S,13S)-9,12-DIOXO-10-(2-BUTYL)-2-OXA-8,11-DIAZABICYCLO [13.2.2] NONADECA-15,17,18-TRIENE] (2R)-BENZYL-(4S)-HYDROXY-5-AMINOPENTANOIC (1R)-HYDROXY-(2S)-INDANEAMIDE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
1N2R
 
 | | A natural selected dimorphism in HLA B*44 alters self, peptide reportoire and T cell recognition. | | Descriptor: | ACETIC ACID, Beta-2-microglobulin, HLA DPA*0201 PEPTIDE, ... | | Authors: | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | Deposit date: | 2002-10-24 | | Release date: | 2004-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
1MW2
 
 | | Amylosucrase soaked with 100mM sucrose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, amylosucrase, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
1MW3
 
 | | Amylosucrase soaked with 1M sucrose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, amylosucrase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
