7D0E
 
 | | Crystal structure of FIP200 Claw/p-CCPG1 FIR2 | | Descriptor: | 3-(2-hydroxyethyloxy)-2-[2-(2-hydroxyethyloxy)ethoxymethyl]-2-(2-hydroxyethyloxymethyl)propan-1-ol, Cell cycle progression protein 1 FIR2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
7CZM
 
 | | Crystal structure of FIP200 Claw/p-OPtineurin LIR complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Optineurin LIR, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
7CZG
 
 | | Crystal structure of FIP200 Claw domain apo form | | Descriptor: | DI(HYDROXYETHYL)ETHER, RB1-inducible coiled-coil protein 1 | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-08 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
3UOT
 
 | | Crystal Structure of MDC1 FHA Domain in Complex with a Phosphorylated Peptide from the MDC1 N-terminus | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Clapperton, J.A, Lloyd, J, Haire, L.F, Li, J, Smerdon, S.J. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator.
Nucleic Acids Res., 40, 2012
|
|
8X8A
 
 | | Crystal structure of STBD1 LIR motif in complex with GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, Starch-binding domain-containing protein 1 | | Authors: | Zhang, Y.C, Pan, L.F. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Decoding the molecular mechanism of selective autophagy of glycogen mediated by autophagy receptor STBD1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5J5T
 
 | | GLK co-crystal structure with aminopyrrolopyrimidine inhibitor | | Descriptor: | 5-[2-(piperidin-4-yl)-1,3-thiazol-5-yl]-3-[(pyridin-4-yl)methoxy]pyridin-2-amine, Mitogen-activated protein kinase kinase kinase kinase 3 | | Authors: | Silvian, L.F, Marcotte, D. | | Deposit date: | 2016-04-03 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Germinal-center kinase-like kinase co-crystal structure reveals a swapped activation loop and C-terminal extension.
Protein Sci., 26, 2017
|
|
3V52
 
 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | 1,2-ETHANEDIOL, ANTI-MHC-I MONOCLONAL ANTIBODY, 64-3-7 H CHAIN, ... | | Authors: | Mage, M.G, Dolan, M.A, Wang, R, Boyd, L.F, Revilleza, M.J, Robinson, H, Natarajan, K, Myers, N.B, Hansen, T.H, Margulies, D.H. | | Deposit date: | 2011-12-15 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
3T8X
 
 | | Crystal structure of human CD1b in complex with synthetic antigenic diacylsulfoglycolipid SGL12 and endogenous spacer | | Descriptor: | 2-O-sulfo-alpha-D-glucopyranosyl 2-O-hexadecanoyl-3-O-[(2E,4S,6S,8S)-2,4,6,8-tetramethyltetracos-2-enoyl]-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Garcia-Alles, L.F, Maveyraud, L, Mourey, L, Julien, S. | | Deposit date: | 2011-08-02 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural reorganization of the human CD1b Antigen-binding groove for presentation of mycobacterial sulfoglycolipids
To be Published
|
|
8X8K
 
 | | Crystal structure of STBD1 CBM20 domain in complex with maltotetraose | | Descriptor: | GLYCEROL, Starch-binding domain-containing protein 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhang, Y.C, Pan, L.F. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decoding the molecular mechanism of selective autophagy of glycogen mediated by autophagy receptor STBD1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7E6Q
 
 | | Crystal structure of influenza A virus neuraminidase N5 complexed with 4'-phenyl-1,2,3-triazolylated oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-acetamido-3-pentan-3-yloxy-5-(4-phenyl-1,2,3-triazol-1-yl)cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, P.F, Babayemi, O.O, Li, C.N, Fu, L.F, Zhang, S.S, Qi, J.X, Lv, X, Li, X.B. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design of 5'-substituted 1,2,3-triazolylated oseltamivir derivatives as potent influenza neuraminidase inhibitors.
Rsc Adv, 11, 2021
|
|
5JOW
 
 | | Bacteroides ovatus Xyloglucan PUL GH43A | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-reducing end alpha-L-arabinofuranosidase BoGH43A | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
7EIN
 
 | | SARS-CoV-2 main proteinase complex with microbial metabolite leupeptin | | Descriptor: | 3C-like proteinase, leupeptin | | Authors: | Fu, L.F, Feng, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Microbial Metabolite Leupeptin in the Treatment of COVID-19 by Traditional Chinese Medicine Herbs.
Mbio, 12, 2021
|
|
7DXM
 
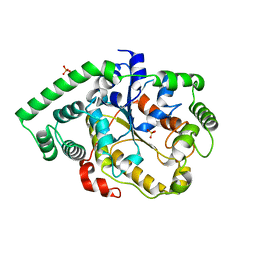 | | Crystal structure of DltD | | Descriptor: | Protein DltD, SULFATE ION | | Authors: | Yan, X.X, Zeng, Q, Tian, L.F. | | Deposit date: | 2021-01-19 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal Structure of an O-acyltransfer Terminal Protein stDltD and Its Implications for dlt Operon-mediated D-alanylation of S. thermophilus.
Prog.Biochem.Biophys., 48, 2022
|
|
1MI2
 
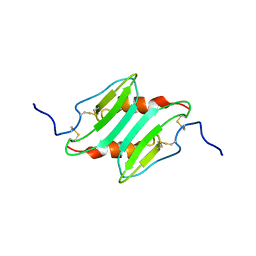 | | SOLUTION STRUCTURE OF MURINE MACROPHAGE INFLAMMATORY PROTEIN-2, NMR, 20 STRUCTURES | | Descriptor: | MACROPHAGE INFLAMMATORY PROTEIN-2 | | Authors: | Shao, W, Jerva, L.F, West, J, Lolis, E, Schweitzer, B.I. | | Deposit date: | 1997-10-24 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of murine macrophage inflammatory protein-2.
Biochemistry, 37, 1998
|
|
1NWM
 
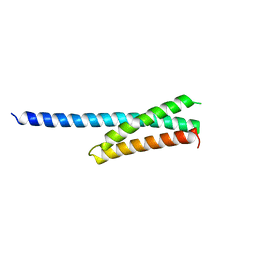 | | GAT domain of human GGA1 | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Suer, S, Misra, S, Saidi, L.F, Hurley, J.H. | | Deposit date: | 2003-02-06 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the GAT domain of human GGA1: a syntaxin amino-terminal domain fold in an endosomal trafficking adaptor.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5M03
 
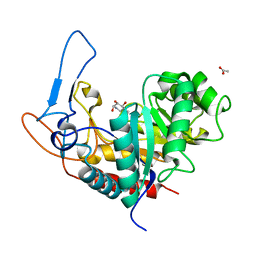 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-noeuromycin and 1,2-alpha-mannobiose | | Descriptor: | (2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
8YSA
 
 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H102 | | Descriptor: | 3C-like proteinase nsp5, BOC-TBG-PHE-ELL | | Authors: | Zheng, W.Y, Fu, L.F, Feng, Y, Han, P, Qi, J.X. | | Deposit date: | 2024-03-22 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery, Biological Activity, and Structural Mechanism of a Potent Inhibitor of SARS-CoV-2 Main Protease
To Be Published
|
|
5M3W
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-1,2-dideoxymannose and alpha-1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
3U1N
 
 | | Structure of the catalytic core of human SAMHD1 | | Descriptor: | PHOSPHATE ION, SAM domain and HD domain-containing protein 1, ZINC ION | | Authors: | Goldstone, D.C, Ennis-Adeniran, V, Walker, P.A, Haire, L.F, Webb, M, Taylor, I.A. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase
Nature, 480, 2011
|
|
5LWP
 
 | | Discovery of phenoxyindazoles and phenylthioindazoles as RORg inverse agonists | | Descriptor: | 4-[3-[2-chloranyl-6-(trifluoromethyl)phenoxy]-5-(dimethylcarbamoyl)indazol-1-yl]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Ouvry, G, Bouix-Peter, C, Ciesielski, F, Chantalat, L, Christin, O, Comino, C, Duvert, D, Feret, C, Harris, C.S, Luzy, A.-P, Musicki, B, Orfila, D, Pascau, J, Parnet, V, Perrin, A, Pierre, R, Raffin, C, Rival, Y, Taquet, N, Thoreau, E, Hennequin, L.F. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of phenoxyindazoles and phenylthioindazoles as ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5M17
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-1,2-dideoxymannose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
1L8G
 
 | | Crystal structure of PTP1B complexed with 7-(1,1-Dioxo-1H-benzo[d]isothiazol-3-yloxymethyl)-2-(oxalyl-amino)-4,7-dihydro-5H-thieno[2,3-c]pyran-3-carboxylic acid | | Descriptor: | 7-(1,1-DIOXO-1H-BENZO[D]ISOTHIAZOL-3-YLOXYMETHYL)-2-(OXALYL-AMINO)-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Iversen, L.F, Andersen, H.S, Moller, K.B, Olsen, O.H, Peters, G.H, Branner, S, Mortensen, S.B, Hansen, T.K, Lau, J, Ge, Y, Holsworth, D.D, Newman, M.J, Moller, N.P.H. | | Deposit date: | 2002-03-20 | | Release date: | 2002-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steric hindrance as a basis for structure-based design of selective inhibitors of protein-tyrosine phosphatases.
Biochemistry, 40, 2001
|
|
3UN0
 
 | | Crystal Structure of MDC1 FHA Domain | | Descriptor: | Mediator of DNA damage checkpoint protein 1, SULFATE ION | | Authors: | Clapperton, J.A, Lloyd, J, Haire, L.F, Li, J, Smerdon, S.J. | | Deposit date: | 2011-11-15 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator.
Nucleic Acids Res., 40, 2012
|
|
3V6L
 
 | | Crystal Structure of caspase-6 inactivation mutation | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.J, Liu, D.F, Li, L.F, Su, X.D. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
J.Biol.Chem., 287, 2012
|
|
3V6M
 
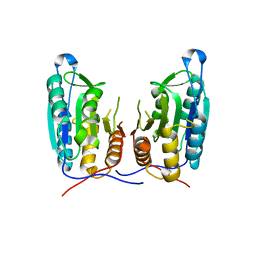 | | Inhibition of caspase-6 activity by single mutation outside the active site | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.J, Liu, D.F, Li, L.F, Su, X.D. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
J.Biol.Chem., 287, 2012
|
|
