4XRQ
 
 | |
8EPU
 
 | |
8EPV
 
 | | 2.2 A crystal structure of the lipocalin cat allergen Fel d 7 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Fel d 7 allergen, ... | | Authors: | Min, J, Pedersen, L.C, Geoffrey, M.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and ligand binding analysis of the pet allergens Can f 1 and Fel d 7.
Front Allergy, 4, 2023
|
|
4B03
 
 | | 6A Electron cryomicroscopy structure of immature Dengue virus serotype 1 | | Descriptor: | DENGUE VIRUS 1 E PROTEIN, DENGUE VIRUS 1 PRM PROTEIN | | Authors: | Kostyuchenko, V.A, Zhang, Q, Tan, L.C, Ng, T.S, Lok, S.M. | | Deposit date: | 2012-06-28 | | Release date: | 2013-06-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Immature and Mature Dengue Serotype 1 Virus Structures Provide Insight Into the Maturation Process.
J.Virol., 87, 2013
|
|
7SCD
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-1 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
7S97
 
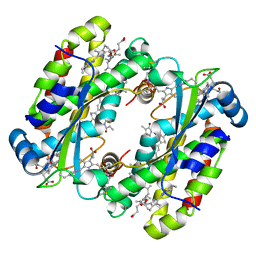 | | Structure of the Photoacclimated Light Harvesting Complex PC577 from Hemiselmis pacifica | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7S96
 
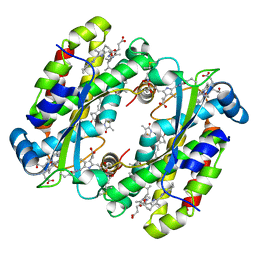 | | Structure of the Light Harvesting Complex PC577 from Hemiselmis pacifica | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7SCE
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-2 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
8D3B
 
 | |
4YOA
 
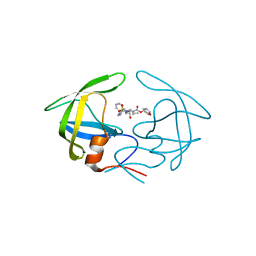 | | Crsystal structure HIV-1 Protease MDR769 L33F Complexed with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 Protease | | Authors: | Kuiper, B.D, Keusch, B, Dewdney, T.G, Chordia, P, Brunzelle, J.S, Ross, K, Kovari, I.A, MacArthur, R, Salimnia, H, Kovari, L.C. | | Deposit date: | 2015-03-11 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | The L33F darunavir resistance mutation acts as a molecular anchor reducing the flexibility of the HIV-1 protease 30s and 80s loops.
Biochem Biophys Rep, 2, 2015
|
|
4YD1
 
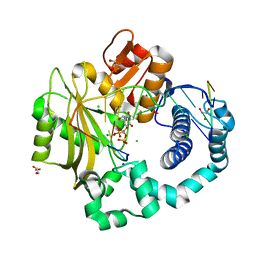 | | Ternary complex of human DNA Polymerase Mu with 2-nt gapped DNA substrate and an incoming nonhydrolyzable dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Moon, A.F, Gosavi, R.A, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2015-02-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Creative template-dependent synthesis by human polymerase mu.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YOB
 
 | | Crystal Structure of Apo HIV-1 Protease MDR769 L33F | | Descriptor: | HIV-1 Protease | | Authors: | Kuiper, B.D, Keusch, B, Dewdney, T.G, Chordia, P, Ross, K, Brunzelle, J.S, Kovari, I.A, MacArthur, R, Salimnia, H, Kovari, L.C. | | Deposit date: | 2015-03-11 | | Release date: | 2015-07-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | The L33F darunavir resistance mutation acts as a molecular anchor reducing the flexibility of the HIV-1 protease 30s and 80s loops.
Biochem Biophys Rep, 2, 2015
|
|
3ZKP
 
 | |
7KJU
 
 | | Cgi121-tRNA complex | | Descriptor: | MAGNESIUM ION, RNA (75-MER) | | Authors: | Ceccarelli, D.F, Beenstock, J, Wan, L.C.K, Sicheri, F. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | A substrate binding model for the KEOPS tRNA modifying complex.
Nat Commun, 11, 2020
|
|
7JYZ
 
 | | Solution NMR structure and dynamics of human Brd3 ET in complex with MLV IN CTD | | Descriptor: | Bromodomain-containing protein 3, Integrase | | Authors: | Aiyer, S, Liu, G, Swapna, G.V.T, Hao, J, Ma, L.C, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-09-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
7KU0
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 138 (yellow) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU1
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 139 (green) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTZ
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 131 (purple) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7BBF
 
 | |
3KKJ
 
 | | X-ray structure of P. syringae q888a4 Oxidoreductase at resolution 2.5A, Northeast Structural Genomics Consortium target PsR10 | | Descriptor: | Amine oxidase, flavin-containing, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Vorobiev, S, Acton, T, Ma, L.C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of P. syringae q888a4 Oxidoreductase at resolution 2.5A, Northeast Structural Genomics Consortium target PsR10
To be Published
|
|
7BH8
 
 | | 3H4-Fab HLA-E-VL9 co-complex | | Descriptor: | 3H4 Fab heavy chain, 3H4 Fab light chain, Beta-2-microglobulin, ... | | Authors: | Walters, L.C, Rozbesky, D. | | Deposit date: | 2021-01-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mouse and human antibodies bind HLA-E-leader peptide complexes and enhance NK cell cytotoxicity.
Commun Biol, 5, 2022
|
|
2I2L
 
 | | X-ray Crystal Structure of Protein yopX from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR411. | | Descriptor: | YopX protein | | Authors: | Vorobiev, S.M, Zhou, W, Seetharaman, J, Forouhar, F, Kuzin, A.A, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-08-16 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the hypothetical protein yopX from Bacillus subtilis
To be Published
|
|
