5N7D
 
 | | MAGI-1 complexed with a RSK1 peptide | | Descriptor: | CALCIUM ION, GLYCEROL, Membrane-associated guanylate kinase, ... | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2017-02-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamic control of RSK complexes by phosphoswitch-based regulation.
FEBS J., 285, 2018
|
|
4IZL
 
 | |
5FDH
 
 | | CRYSTAL STRUCTURE OF OXA-405 BETA-LACTAMASE | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Retailleau, P, Oueslati, S, Marchini, L, Dortet, L, Naas, T, Iorga, B. | | Deposit date: | 2015-12-16 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Biochemical and Structural Characterization of OXA-405, an OXA-48 Variant with Extended-Spectrum beta-Lactamase Activity.
Microorganisms, 8, 2019
|
|
2BUK
 
 | | SATELLITE TOBACCO NECROSIS VIRUS | | Descriptor: | CALCIUM ION, COAT PROTEIN | | Authors: | Jones, T.A, Liljas, L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of Satellite Tobacco Necrosis Virus After Crystallographic Refinement at 2.5 A Resolution.
J.Mol.Biol., 177, 1984
|
|
5FOJ
 
 | | Cryo electron microscopy structure of Grapevine Fanleaf Virus complex with Nanobody | | Descriptor: | Nanobody, RNA2 polyprotein | | Authors: | Orlov, I, Hemmer, C, Ackerer, L, Lorber, B, Ghannam, A, Poignavent, V, Hleibieh, K, Sauter, C, Schmitt-Keichinger, C, Belval, L, Hily, J.M, Marmonier, A, Komar, V, Gersch, S, Schellenberger, P, Bron, P, Vigne, E, Muyldermans, S, Lemaire, O, Demangeat, G, Ritzenthaler, C, Klaholz, B.P. | | Deposit date: | 2015-11-22 | | Release date: | 2016-01-20 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of nanobody recognition of grapevine fanleaf virus and of virus resistance loss.
Proc.Natl.Acad.Sci.USA, 2020
|
|
5HXG
 
 | | STM1697-FlhD complex | | Descriptor: | Flagellar transcriptional regulator FlhD, Uncharacterized protein STM1697 | | Authors: | Li, B, Yuan, Z, Qin, L, Gu, L. | | Deposit date: | 2016-01-30 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of STM1697-FlhD complex
To Be Published
|
|
1M8R
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 7.4) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase A2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
3BID
 
 | | Crystal structure of the NMB1088 protein from Neisseria meningitidis. Northeast Structural Genomics Consortium target MR91 | | Descriptor: | UPF0339 protein NMB1088 | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, L, Fang, Y, Xiao, R, Owen, L.A, Maglaqui, M, Cunningham, K, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the NMB1088 protein from Neisseria meningitidis.
To be Published
|
|
3BB6
 
 | | Crystal structure of the P64488 protein from E.coli (strain K12). Northeast Structural Genomics Consortium target ER596 | | Descriptor: | Uncharacterized protein yeaR, ZINC ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Janjua, H, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray structure of the P64488 from E.coli (strain K12).
To be Published
|
|
5MSZ
 
 | | Lytic Polysaccharide Monooxygenase AA15 from Thermobia domestica in the Cu(I) State | | Descriptor: | 1,2-ETHANEDIOL, COPPER (I) ION, Thermobia domestica domestica AA15 | | Authors: | Hemsworth, G.R, Sabbadin, F, Ciano, L, Henrissat, B, Dupree, P, Tryfona, T, Besser, K, Elias, L, Pesante, G, Li, Y, Dowle, A, Bates, R, Gomez, L, Hallam, R, Davies, G.J, Walton, P.H, Bruce, N.C, McQueen-Mason, S. | | Deposit date: | 2017-01-06 | | Release date: | 2018-02-28 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion.
Nat Commun, 9, 2018
|
|
6ENM
 
 | | Crystal structure of MMP12 in complex with hydroxamate inhibitor LP168. | | Descriptor: | 2-[2-[4-(4-methoxyphenyl)phenyl]sulfonylphenyl]-~{N}-oxidanyl-ethanamide, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Vera, L, Nuti, E, Rossello, A, Stura, E.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of Thioaryl-Based Matrix Metalloproteinase-12 Inhibitors with Alternative Zinc-Binding Groups: Synthesis, Potentiometric, NMR, and Crystallographic Studies.
J. Med. Chem., 61, 2018
|
|
5EY9
 
 | | Structure of FadD32 from Mycobacterium marinum complexed to AMPC12 | | Descriptor: | GLYCEROL, Long-chain-fatty-acid--AMP ligase FadD32, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl icosyl hydrogen phosphate | | Authors: | Guillet, V, Maveyraud, L, Mourey, L. | | Deposit date: | 2015-11-24 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into Structure-Function Relationships and Inhibition of the Fatty Acyl-AMP Ligase (FadD32) Orthologs from Mycobacteria.
J.Biol.Chem., 291, 2016
|
|
1O9Z
 
 | | F17-aG lectin domain from Escherichia coli (ligand free) | | Descriptor: | F17-AG LECTIN DOMAIN | | Authors: | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2002-12-23 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
5NXQ
 
 | |
6XX1
 
 | | The unique CBM3-Clocl_1192 of Hungateiclostridium clariflavum | | Descriptor: | Cellulose binding domain-containing protein | | Authors: | Milana, M.V, Almog, R, Yaniv, O, Oded, L, Inna, R.G, Felix, F, Edward, A.B, Raphael, L. | | Deposit date: | 2020-01-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The unique CBM3-Cthe_0271 from Ruminoclostridium thermocellum and CBM3-Clocl_1192 from Hungateiclostridium clariflavum
To Be Published
|
|
1M8S
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 5.9) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase a2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
2EB2
 
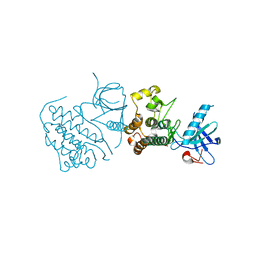 | | Crystal structure of mutated EGFR kinase domain (G719S) | | Descriptor: | Epidermal growth factor receptor | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Senba, K, Yamamoto, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-06 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 2012
|
|
6XUH
 
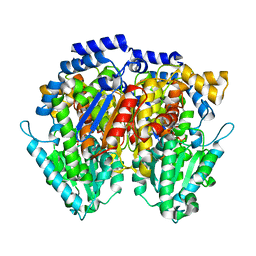 | | Crystal structure of human phosphoglucose isomerase in complex with inhibitor | | Descriptor: | (2R,3R,4S)-5-((2-aminoethyl)amino)-2,3,4-trihydroxy-5-oxopentyl dihydrogen phosphate, 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase | | Authors: | Li de la Sierra-Gallay, I, Ahmad, L, Plancqueel, S, van Tilbeurgh, H, Salmon, L. | | Deposit date: | 2020-01-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Novel N-substituted 5-phosphate-d-arabinonamide derivatives as strong inhibitors of phosphoglucose isomerases: Synthesis, structure-activity relationship and crystallographic studies.
Bioorg.Chem., 102, 2020
|
|
1KQK
 
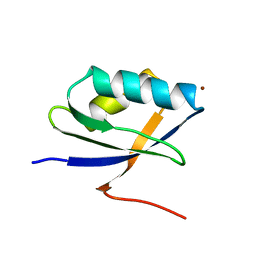 | | Solution Structure of the N-terminal Domain of a Potential Copper-translocating P-type ATPase from Bacillus subtilis in the Cu(I)loaded State | | Descriptor: | COPPER (I) ION, POTENTIAL COPPER-TRANSPORTING ATPASE | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F.C, Ruiz-Duenas, F.J. | | Deposit date: | 2002-01-07 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
8B56
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GD-9 | | Descriptor: | (2~{S})-4-(2-chloranylethanoyl)-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5, BROMIDE ION, ... | | Authors: | Straeter, N, Muller, C.E, Claff, T, Sylvester, K, Weisse, R, Gao, S, Song, L, Liu, X, Zhan, P. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Discovery and Crystallographic Studies of Nonpeptidic Piperazine Derivatives as Covalent SARS-CoV-2 Main Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
1KV2
 
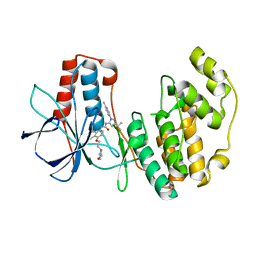 | | Human p38 MAP Kinase in Complex with BIRB 796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, p38 MAP kinase | | Authors: | Pargellis, C, Tong, L, Churchill, L, Cirillo, P.F, Gilmore, T, Graham, A.G, Grob, P.M, Hickey, E.R, Moss, N, Pav, S, Regan, J. | | Deposit date: | 2002-01-23 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site.
Nat.Struct.Biol., 9, 2002
|
|
5HD4
 
 | | Dissecting Therapeutic Resistance to ERK Inhibition Rat Wild Type SCH772984 in complex with (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide | | Descriptor: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Jha, S, Morris, E.J, Hruza, A, Mansueto, M.S, Schroeder, G, Arbanas, J, McMasters, D, Restaino, C.R, Dayananth, R, Black, S, Elsen, N.L, Mannarino, A, Cooper, A, Fawell, S, Zawel, L, Jayaraman, L, Samatar, A.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dissecting Therapeutic Resistance to ERK Inhibition.
Mol.Cancer Ther., 15, 2016
|
|
1KV1
 
 | | p38 MAP Kinase in Complex with Inhibitor 1 | | Descriptor: | 1-(5-TERT-BUTYL-2-METHYL-2H-PYRAZOL-3-YL)-3-(4-CHLORO-PHENYL)-UREA, p38 MAP kinase | | Authors: | Pargellis, C, Tong, L, Churchill, L, Cirillo, P.F, Gilmore, T, Graham, A.G, Grob, P.M, Hickey, E.R, Moss, N, Pav, S, Regan, J. | | Deposit date: | 2002-01-23 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site.
Nat.Struct.Biol., 9, 2002
|
|
5N7G
 
 | | MAGI-1 complexed with a synthetic pRSK1 peptide | | Descriptor: | CALCIUM ION, GLYCEROL, Membrane-associated guanylate kinase, ... | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2017-02-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dynamic control of RSK complexes by phosphoswitch-based regulation.
FEBS J., 285, 2018
|
|
8C1V
 
 | | SARS-CoV-2 S-trimer (3 RBDs up) bound to TriSb92, fitted into cryo-EM map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sb92, ... | | Authors: | Huiskonen, J.T, Rissanen, I, Hannula, L. | | Deposit date: | 2022-12-21 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Intranasal trimeric sherpabody inhibits SARS-CoV-2 including recent immunoevasive Omicron subvariants.
Nat Commun, 14, 2023
|
|
