1MST
 
 | |
6JUF
 
 | | SspB crystal structure | | Descriptor: | SspB protein | | Authors: | Liqiong, L, Yubing, Z. | | Deposit date: | 2019-04-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
5GOK
 
 | | K11/K63-branched tri-Ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GP7
 
 | | Structural basis for the binding between Tankyrase-1 and USP25 | | Descriptor: | GLYCEROL, Tankyrase-1, Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Liu, J, Xu, D, Fu, T, Pan, L. | | Deposit date: | 2016-08-01 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | USP25 regulates Wnt signaling by controlling the stability of tankyrases
Genes Dev., 31, 2017
|
|
2ZKS
 
 | | Structural insights into the proteolytic machinery of apoptosis-inducing Granzyme M | | Descriptor: | Granzyme M, SULFATE ION, hGzmM inhibitor | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Yang, X, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-03-28 | | Release date: | 2009-03-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
5F7E
 
 | | Crystal structure of germ-line precursor of 3BNC60 Fab | | Descriptor: | Fab heavy chain, Fab light chain | | Authors: | Sievers, S.A, Scharf, L, Jiang, S, Bjorkman, P.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
6FXY
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Gal - Structure from long-wavelength S-SAD | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
5TBI
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1427 | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-[2-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylamino]ethylamino]-1-(methoxymethyl)pyrimidin-2-one, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
2AAA
 
 | | CALCIUM BINDING IN ALPHA-AMYLASES: AN X-RAY DIFFRACTION STUDY AT 2.1 ANGSTROMS RESOLUTION OF TWO ENZYMES FROM ASPERGILLUS | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z, Dodson, E.J, Dodson, G.G. | | Deposit date: | 1991-02-27 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Calcium binding in alpha-amylases: an X-ray diffraction study at 2.1-A resolution of two enzymes from Aspergillus.
Biochemistry, 29, 1990
|
|
5I0K
 
 | |
1KTI
 
 | | BINDING OF 100 MM N-ACETYL-N'-BETA-D-GLUCOPYRANOSYL UREA TO GLYCOGEN PHOSPHORYLASE B: KINETIC AND CRYSTALLOGRAPHIC STUDIES | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-(acetylcarbamoyl)-beta-D-glucopyranosylamine, ... | | Authors: | Oikonomakos, N.G, Kosmopoulou, M, Zographos, S.E, Leonidas, D.D, Chrysina, E.D, Somsak, L, Nagy, V, Praly, J.P, Docsa, T, Toth, B, Gergely, P. | | Deposit date: | 2002-01-16 | | Release date: | 2002-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Binding of N-acetyl-N'-beta-D-glucopyranosyl urea and N-benzoyl-N'-beta-D-glucopyranosyl urea to glycogen phosphorylase b: kinetic and crystallographic studies.
Eur.J.Biochem., 269, 2002
|
|
6FXM
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Mn2+ | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
3TGL
 
 | | STRUCTURE AND MOLECULAR MODEL REFINEMENT OF RHIZOMUCOR MIEHEI TRIACYLGLYCERIDE LIPASE: A CASE STUDY OF THE USE OF SIMULATED ANNEALING IN PARTIAL MODEL REFINEMENT | | Descriptor: | TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z.S, Dodson, E.J, Dodson, G.G, Tolley, S.P, Turkenburg, J.P, Christiansen, L, Huge-Jensen, B, Norskov, L, Thim, L. | | Deposit date: | 1991-07-29 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURE AND MOLECULAR-MODEL REFINEMENT OF RHIZOMUCOR-MIEHEI TRIACYLGLYCERIDE LIPASE - A CASE-STUDY OF THE USE OF SIMULATED ANNEALING IN PARTIAL MODEL REFINEMENT.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
4FZO
 
 | |
4FZP
 
 | |
1N86
 
 | | Crystal structure of human D-dimer from cross-linked fibrin complexed with GPR and GHRPLDK peptide ligands. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CALCIUM ION, Fibrin alpha/alpha-E chain, ... | | Authors: | Yang, Z, Pandi, L, Doolittle, R.F. | | Deposit date: | 2002-11-19 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of fragment double-D from cross-linked lamprey fibrin reveals isopeptide linkages across an unexpected D-D interface.
Biochemistry, 41, 2002
|
|
1ND1
 
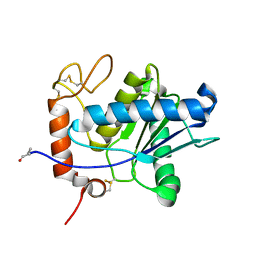 | | Amino acid sequence and crystal structure of BaP1, a metalloproteinase from Bothrops asper snake venom that exerts multiple tissue-damaging activities. | | Descriptor: | BaP1, ZINC ION | | Authors: | Watanabe, L, Shannon, J.D, Valente, R.H, Rucavado, A, Alape-Giron, A, Kamiguti, A.S, Theakston, R.D, Fox, J.W, Gutierrez, J.M, Arni, R.K. | | Deposit date: | 2002-12-06 | | Release date: | 2003-11-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Amino acid sequence and crystal structure of BaP1, a metalloproteinase from Bothrops asper snake venom that exerts multiple tissue-damaging activities
Protein Sci., 12, 2003
|
|
3CF8
 
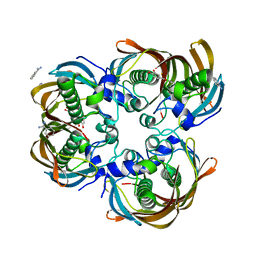 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with quercetin | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 3,5,7,3',4'-PENTAHYDROXYFLAVONE, BENZAMIDINE, ... | | Authors: | Zhang, L, Wu, D, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2008-03-03 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: crystal structure characterization with enzymatic inhibition assay
Protein Sci., 17, 2008
|
|
3CF9
 
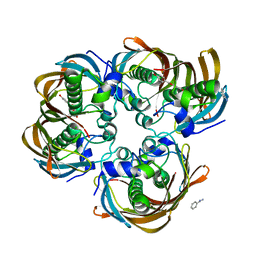 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with apigenin | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, BENZAMIDINE, ... | | Authors: | Zhang, L, Wu, D, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2008-03-03 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: crystal structure characterization with enzymatic inhibition assay
Protein Sci., 17, 2008
|
|
3CX3
 
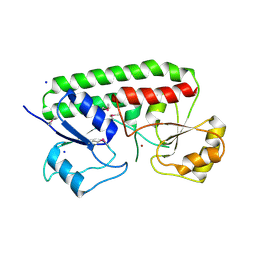 | | Crystal structure Analysis of the Streptococcus pneumoniae AdcAII protein | | Descriptor: | Lipoprotein, SODIUM ION, ZINC ION | | Authors: | Loisel, E, Durmort, C, Jacquamet, L. | | Deposit date: | 2008-04-23 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AdcAII, a new pneumococcal Zn-binding protein homologous with ABC transporters: biochemical and structural analysis.
J.Mol.Biol., 381, 2008
|
|
5LV2
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1246 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
5A7Y
 
 | | Crystal structure of Sulfolobus acidocaldarius Trm10 in complex with S-adenosylhomocysteine | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | Deposit date: | 2015-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
6CSV
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 63 kDa,Centrosomal protein of 152 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
7Y42
 
 | |
7XTQ
 
 | | Cryo-EM structure of the R399-bound GPBAR-Gs complex | | Descriptor: | G-protein coupled bile acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, L, Yang, F, Wu, X, Mao, C, Sun, J, Yu, X, Zhang, Y, Zhang, P. | | Deposit date: | 2022-05-17 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis and molecular mechanism of biased GPBAR signaling in regulating NSCLC cell growth via YAP activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
