3B3N
 
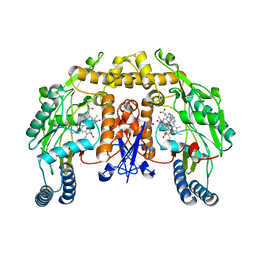 | | Structure of neuronal NOS heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-aminopyridin-2"-yl)methyl]pyrrolidin-3'-yl}ethane-1,2-diamine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-aminopyridin-2-yl)methyl]pyrrolidin-3-yl}ethane-1,2-diamine, ... | | 著者 | Igarashi, J, Li, H, Poulos, T.L. | | 登録日 | 2007-10-22 | | 公開日 | 2008-07-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Minimal pharmacophoric elements and fragment hopping, an approach directed at molecular diversity and isozyme selectivity. Design of selective neuronal nitric oxide synthase inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
6OI9
 
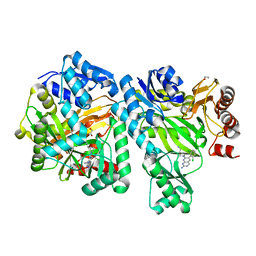 | | Crystal Structure of E. coli Biotin Carboxylase Complexed with 7-[3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine | | 分子名称: | 1,2-ETHANEDIOL, 7-[(3S)-3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | 著者 | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | 登録日 | 2019-04-09 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
2PEG
 
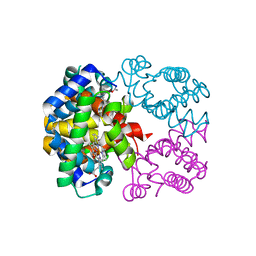 | | Crystal structure of Trematomus bernacchii hemoglobin in a partial hemichrome state | | 分子名称: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Vergara, A, Franzese, M, Merlino, A, Vitagliano, L, Mazzarella, L. | | 登録日 | 2007-04-03 | | 公開日 | 2007-07-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Structural characterization of ferric hemoglobins from three antarctic fish species of the suborder notothenioidei.
Biophys.J., 93, 2007
|
|
6OPO
 
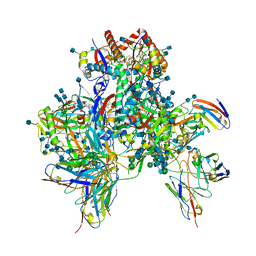 | | Symmetric model of CD4- and 17-bound B41 HIV-1 Env SOSIP in complex with DDM | | 分子名称: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ozorowski, G, Torres, J.L, Ward, A.B. | | 登録日 | 2019-04-25 | | 公開日 | 2020-10-21 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
7QNM
 
 | | Crystallization and structural analyses of ZgHAD, a L-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans | | 分子名称: | (S)-2-haloacid dehalogenase, PHOSPHATE ION | | 著者 | Grigorian, E, Roret, T, Leblanc, C, Delage, L, Czjzek, M. | | 登録日 | 2021-12-21 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | X-ray structure and mechanism of ZgHAD, a l-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans.
Protein Sci., 32, 2023
|
|
6O7X
 
 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 3 | | 分子名称: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | 登録日 | 2019-03-08 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (8.7 Å) | | 主引用文献 | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6OCP
 
 | | Crystal structure of a human GABAB receptor peptide bound to KCTD16 T1 | | 分子名称: | BTB/POZ domain-containing protein KCTD16, Gamma-aminobutyric acid type B receptor subunit 2 | | 著者 | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | 登録日 | 2019-03-25 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3B8W
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221P | | 分子名称: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | 登録日 | 2007-11-02 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
4M1L
 
 | | Complex of IQCG and Ca2+-bound CaM | | 分子名称: | CALCIUM ION, Calmodulin, IQ domain-containing protein G, ... | | 著者 | Liang, W.X, Chen, L.T, Chen, Z, Chen, S.J, Chen, S. | | 登録日 | 2013-08-03 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Functional and molecular features of the calmodulin-interacting protein IQCG required for haematopoiesis in zebrafish
Nat Commun, 5, 2014
|
|
4M27
 
 | | Crystal structure of non-heme iron oxygenase OrfP in complex with Fe and L-Arg | | 分子名称: | ARGININE, FE (III) ION, L-arginine beta-hydroxylase, ... | | 著者 | Chang, C.Y, Liu, Y.C, Lyu, S.Y, Wu, C.C, Li, T.L. | | 登録日 | 2013-08-05 | | 公開日 | 2014-06-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Biosynthesis of streptolidine involved two unexpected intermediates produced by a dihydroxylase and a cyclase through unusual mechanisms.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3B3M
 
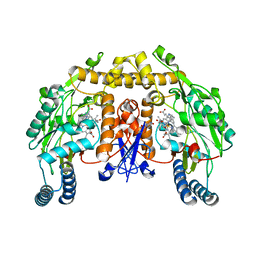 | | Structure of neuronal NOS heme domain in complex with a inhibitor (+-)-3-{cis-4'-[(6"-aminopyridin-2"-yl)methyl]pyrrolidin-3'-ylamino}propan-1-ol | | 分子名称: | 3-({(3S,4S)-4-[(6-aminopyridin-2-yl)methyl]pyrrolidin-3-yl}amino)propan-1-ol, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | 著者 | Igarashi, J, Li, H, Poulos, T.L. | | 登録日 | 2007-10-22 | | 公開日 | 2008-07-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Minimal pharmacophoric elements and fragment hopping, an approach directed at molecular diversity and isozyme selectivity. Design of selective neuronal nitric oxide synthase inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
2PNE
 
 | | Crystal Structure of the Snow Flea Antifreeze Protein | | 分子名称: | 6.5 kDa glycine-rich antifreeze protein | | 著者 | Pentelute, B.L, Kent, S.B.H, Gates, Z.P, Tereshko, V, Kossiakoff, A.A, Kurutz, J, Dashnau, J, Vaderkooi, J.M. | | 登録日 | 2007-04-24 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers
J.Am.Chem.Soc., 130, 2008
|
|
3B5I
 
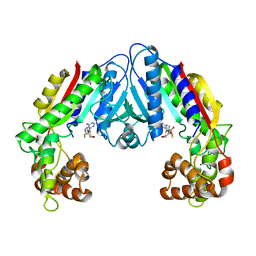 | | Crystal structure of Indole-3-acetic Acid Methyltransferase | | 分子名称: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase-like protein | | 著者 | Ferrer, J.-L, Noel, J.P. | | 登録日 | 2007-10-26 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural, Biochemical, and Phylogenetic Analyses Suggest That Indole-3-Acetic Acid Methyltransferase Is an Evolutionarily Ancient Member of the SABATH Family.
Plant Physiol., 146, 2008
|
|
6EQP
 
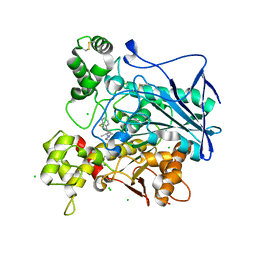 | | Human butyrylcholinesterase in complex with ethopropazine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Nachon, F, Brazzolotto, X, Wandhammer, M, Trovaslet-Leroy, M, Rosenberry, T.L, Macdonald, I.R, Darvesh, S. | | 登録日 | 2017-10-14 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.349417 Å) | | 主引用文献 | Comparison of the Binding of Reversible Inhibitors to Human Butyrylcholinesterase and Acetylcholinesterase: A Crystallographic, Kinetic and Calorimetric Study.
Molecules, 22, 2017
|
|
8DRW
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp9-nsp10 (C9) cut site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp9-nsp10 (C9) cut site, PENTAETHYLENE GLYCOL, ... | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8J5K
 
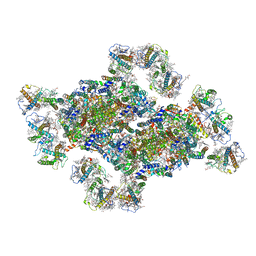 | | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana | | 分子名称: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | 著者 | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | 登録日 | 2023-04-23 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
8DRY
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp12-nsp13 (C12) cut site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp12-nsp13 (C12) cut site | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
6OO7
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in nanodiscs | | 分子名称: | TRPV2, resiniferatoxin | | 著者 | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | 登録日 | 2019-04-22 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
3B2X
 
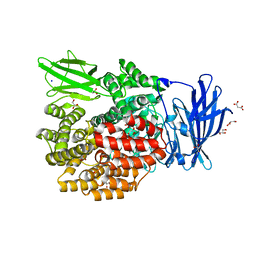 | | Crystal Structure of E. coli Aminopeptidase N in complex with Lysine | | 分子名称: | Aminopeptidase N, GLYCEROL, LYSINE, ... | | 著者 | Addlagatta, A, Gay, L, Matthews, B.W. | | 登録日 | 2007-10-19 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis for the unusual specificity of Escherichia coli aminopeptidase N.
Biochemistry, 47, 2008
|
|
6OPQ
 
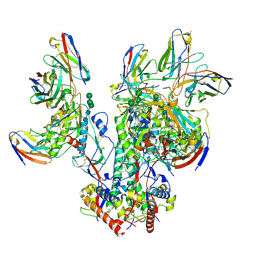 | | CD4- and 17-bound HIV-1 Env B41 SOSIP frozen with LMNG | | 分子名称: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ozorowski, G, Torres, J.L, Ward, A.B. | | 登録日 | 2019-04-25 | | 公開日 | 2020-10-21 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
6OS0
 
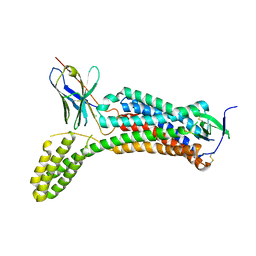 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to angiotensin II | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensinogen, CHLORIDE ION, ... | | 著者 | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | 登録日 | 2019-05-01 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
6OY4
 
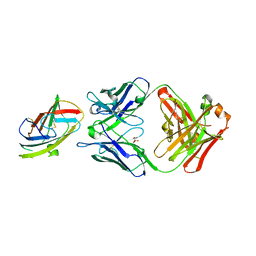 | | Crystal structure of complex between recombinant Der p 2.0103 and Fab fragment of 7A1 | | 分子名称: | Der p 2 variant 3, Fab fragment of IgG, HEAVY CHAIN, ... | | 著者 | Kapingidza, A.B, Offermann, L.R, Glesner, J, Wunschmann, S, Vailes, L.D, Chapman, M.D.C, Pomes, A, Chruszcz, M. | | 登録日 | 2019-05-14 | | 公開日 | 2019-08-28 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | A Human IgE Antibody Binding Site on Der p 2 for the Design of a Recombinant Allergen for Immunotherapy.
J Immunol., 203, 2019
|
|
3B8T
 
 | | Crystal structure of Escherichia coli alaine racemase mutant P219A | | 分子名称: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | 登録日 | 2007-11-02 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
2PJQ
 
 | | Crystal structure of Q88U62_LACPL from Lactobacillus plantarum. Northeast Structural Genomics target LpR71 | | 分子名称: | Uncharacterized protein lp_2664 | | 著者 | Benach, J, Su, M, Seetharaman, J, Forouhar, F, Chen, C.X, Cunningham, K, Ma, L.-C, Owens, L, Baran, M, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-04-16 | | 公開日 | 2007-05-01 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of Q88U62_LACPL from Lactobacillus plantarum.
To be Published
|
|
6EG0
 
 | | Crystal structure of Dpr4 Ig1-Ig2 in complex with DIP-Eta Ig1-Ig3 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Cosmanescu, F, Shapiro, L. | | 登録日 | 2018-08-17 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins.
Neuron, 100, 2018
|
|
