5Y0C
 
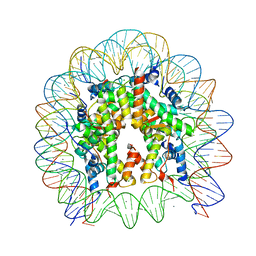 | | Crystal Structure of the human nucleosome at 2.09 angstrom resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | Deposit date: | 2017-07-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
5Y0D
 
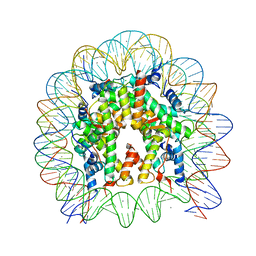 | | Crystal Structure of the human nucleosome containing the H2B E76K mutant | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | Deposit date: | 2017-07-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
6AAV
 
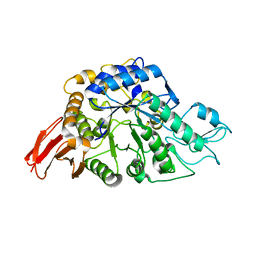 | | Crystal structure of alpha-glucosyl transfer enzyme, XgtA at 1.72 angstrom resolution | | Descriptor: | Alpha-glucosyltransferase | | Authors: | Kurumizaka, H, Arimura, Y, Kirimura, K, Watanabe, R. | | Deposit date: | 2018-07-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of alpha-glucosyl transfer enzyme XgtA from Xanthomonas campestris WU-9701.
Biochem.Biophys.Res.Commun., 526, 2020
|
|
7YOZ
 
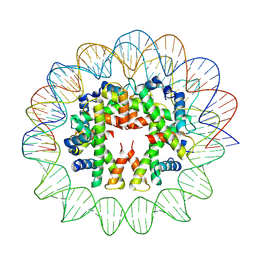 | | Cryo-EM structure of human subnucleosome (intermediate form) | | Descriptor: | Histone H3.1, Histone H4, Widom601 DNA FW (145-MER), ... | | Authors: | Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron microscopy structure of the H3-H4 octasome: A nucleosome-like particle without histones H2A and H2B.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4WD3
 
 | | Crystal structure of an L-amino acid ligase RizA | | Descriptor: | L-amino acid ligase | | Authors: | Kagawa, W, Arai, T, Kino, K, Kurumizaka, H. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of RizA, an L-amino-acid ligase from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3VH0
 
 | | Crystal structure of E. coli YncE complexed with DNA | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*TP*AP*CP*TP*CP*AP*G)-3'), Uncharacterized protein YncE | | Authors: | Kagawa, W, Sagawa, T, Niki, H, Kurumizaka, H. | | Deposit date: | 2011-08-23 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the DNA-binding activity of the bacterial beta-propeller protein YncE
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3VGZ
 
 | | Crystal structure of E. coli YncE | | Descriptor: | Uncharacterized protein YncE | | Authors: | Kagawa, W, Sagawa, T, Niki, H, Kurumizaka, H. | | Deposit date: | 2011-08-23 | | Release date: | 2011-11-16 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the DNA-binding activity of the bacterial beta-propeller protein YncE
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6L4A
 
 | | H3-H3-H3 tri-nucleosome with the 22 base-pair linker DNA | | Descriptor: | DNA (485-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Takizawa, Y, Ho, C.-H, Tachiwana, H, Matsunami, H, Ohi, M, Wolf, M, Kurumizaka, H. | | Deposit date: | 2019-10-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Cryo-EM Structures of Centromeric Tri-nucleosomes Containing a Central CENP-A Nucleosome.
Structure, 28, 2020
|
|
6KVD
 
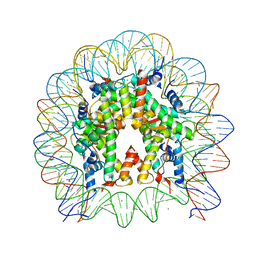 | | Crystal structure of human nucleosome containing H2A.J | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.J, ... | | Authors: | Tanaka, H, Koyama, M, Sato, S, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Biochemical and structural analyses of the nucleosome containing human histone H2A.J.
J.Biochem., 167, 2020
|
|
6A5O
 
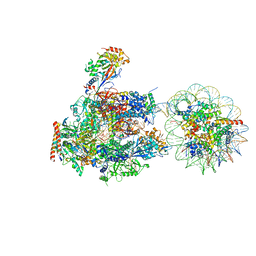 | | RNA polymerase II elongation complex stalled at SHL(-6) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6V2K
 
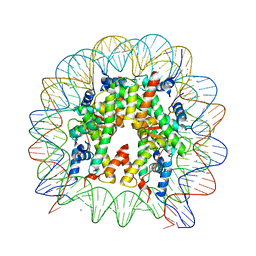 | | The nucleosome structure after H2A-H2B exchange | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A, ... | | Authors: | Arimura, Y, Hirano, R, Kurumizaka, H. | | Deposit date: | 2019-11-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Histone variant H2A.B-H2B dimers are spontaneously exchanged with canonical H2A-H2B in the nucleosome.
Commun Biol, 4, 2021
|
|
7D69
 
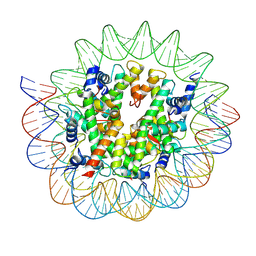 | | Cryo-EM structure of the nucleosome containing Giardia histones | | Descriptor: | 601L DNA (145-MER), Histone H2A, Histone H2B, ... | | Authors: | Sato, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Cryo-EM structure of the nucleosome core particle containing Giardia lamblia histones.
Nucleic Acids Res., 49, 2021
|
|
7DBH
 
 | | The mouse nucleosome structure containing H3mm18 | | Descriptor: | DNA (126-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Hirai, S, Takizawa, Y, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2020-10-20 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant.
Nucleic Acids Res., 50, 2022
|
|
7D20
 
 | | Cryo-EM structure of SET8-CENP-A-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2020-09-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
7D1Z
 
 | | Cryo-EM structure of SET8-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2020-09-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
1HNR
 
 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
1HNS
 
 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
4Z5T
 
 | | The nucleosome containing human H3.5 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Urahama, T, Harada, A, Maehara, K, Horikoshi, N, Sato, K, Sato, Y, Shiraishi, K, Sugino, N, Osakabe, A, Tachiwana, H, Kagawa, W, Kimura, H, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-04-03 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Histone H3.5 forms an unstable nucleosome and accumulates around transcription start sites in human testis.
Epigenetics Chromatin, 9, 2016
|
|
8H0W
 
 | | RNA polymerase II transcribing a chromatosome (type II) | | Descriptor: | DNA (261-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hirano, R, Ehara, H, Tomoya, K, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2022-09-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of RNA polymerase II transcription on the chromatosome containing linker histone H1.
Nat Commun, 13, 2022
|
|
8H0V
 
 | | RNA polymerase II transcribing a chromatosome (type I) | | Descriptor: | DNA (261-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hirano, R, Ehara, H, Tomoya, K, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2022-09-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of RNA polymerase II transcription on the chromatosome containing linker histone H1.
Nat Commun, 13, 2022
|
|
8HE5
 
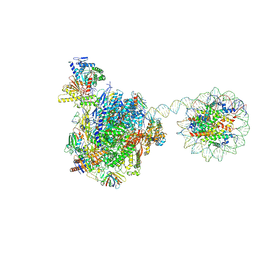 | | RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA repair protein, DNA-directed RNA polymerase subunit, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Kinoshita, C, Saotome, M, Kagawa, W, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.95 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
2EO0
 
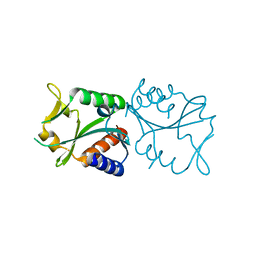 | |
1AA3
 
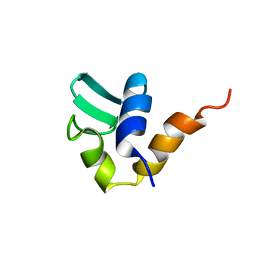 | | C-TERMINAL DOMAIN OF THE E. COLI RECA, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RECA | | Authors: | Aihara, H, Ito, Y, Kurumizaka, H, Terada, T, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-01-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | An interaction between a specified surface of the C-terminal domain of RecA protein and double-stranded DNA for homologous pairing.
J.Mol.Biol., 274, 1997
|
|
1B22
 
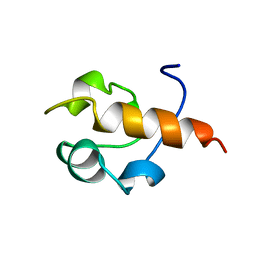 | | RAD51 (N-TERMINAL DOMAIN) | | Descriptor: | DNA REPAIR PROTEIN RAD51 | | Authors: | Aihara, H, Ito, Y, Kurumizaka, H, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR.
J.Mol.Biol., 290, 1999
|
|
7C0M
 
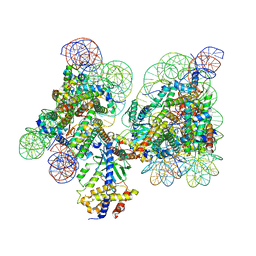 | | Human cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Kujirai, T, Zierhut, C, Takizawa, Y, Kim, R, Negishi, L, Uruma, N, Hirai, S, Funabiki, H, Kurumizaka, H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for the inhibition of cGAS by nucleosomes.
Science, 370, 2020
|
|
