3DQW
 
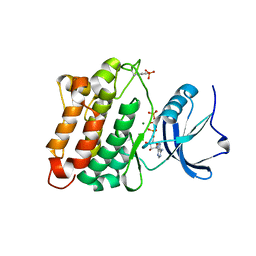 | | c-Src kinase domain Thr338Ile mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Azam, M, Seeliger, M.A, Gray, N, Kuriyan, J, Daley, G.Q. | | Deposit date: | 2008-07-09 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | Activation of tyrosine kinases by mutation of the gatekeeper threonine.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1GIR
 
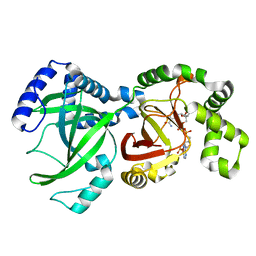 | | CRYSTAL STRUCTURE OF THE ENZYMATIC COMPONET OF IOTA-TOXIN FROM CLOSTRIDIUM PERFRINGENS WITH NADPH | | Descriptor: | IOTA TOXIN COMPONENT IA, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
1A5T
 
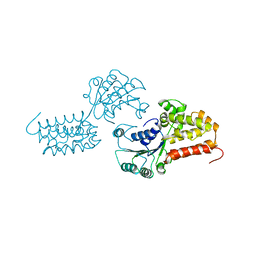 | | CRYSTAL STRUCTURE OF THE DELTA PRIME SUBUNIT OF THE CLAMP-LOADER COMPLEX OF ESCHERICHIA COLI DNA POLYMERASE III | | Descriptor: | DELTA PRIME, ZINC ION | | Authors: | Guenther, B, Onrust, R, Sali, A, O'Donnell, M, Kuriyan, J. | | Deposit date: | 1998-02-18 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the delta' subunit of the clamp-loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 91, 1997
|
|
1EQN
 
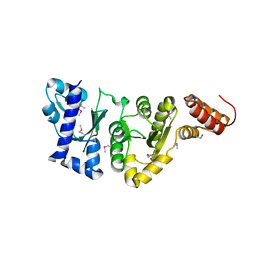 | | E.COLI PRIMASE CATALYTIC CORE | | Descriptor: | DNA PRIMASE | | Authors: | Podobnik, M, McInerney, P, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2000-04-05 | | Release date: | 2000-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases.
J.Mol.Biol., 300, 2000
|
|
1AD5
 
 | | SRC FAMILY KINASE HCK-AMP-PNP COMPLEX | | Descriptor: | CALCIUM ION, HAEMATOPOETIC CELL KINASE HCK, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Sicheri, F, Moarefi, I, Kuriyan, J. | | Deposit date: | 1997-02-20 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the Src family tyrosine kinase Hck.
Nature, 385, 1997
|
|
1BKD
 
 | | COMPLEX OF HUMAN H-RAS WITH HUMAN SOS-1 | | Descriptor: | H-RAS, SON OF SEVENLESS-1 | | Authors: | Boriack-Sjodin, P.A, Margarit, S.M, Sagi, D.B, Kuriyan, J. | | Deposit date: | 1998-07-06 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of the activation of Ras by Sos.
Nature, 394, 1998
|
|
1BK6
 
 | | KARYOPHERIN ALPHA (YEAST) + SV40 T ANTIGEN NLS | | Descriptor: | KARYOPHERIN ALPHA, LARGE T ANTIGEN | | Authors: | Conti, E, Uy, M, Leighton, L, Blobel, G, Kuriyan, J. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha.
Cell(Cambridge,Mass.), 94, 1998
|
|
1GIQ
 
 | | Crystal Structure of the Enzymatic Componet of Iota-Toxin from Clostridium Perfringens with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, IOTA TOXIN COMPONENT IA | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
1BK5
 
 | | KARYOPHERIN ALPHA FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | COBALT (II) ION, KARYOPHERIN ALPHA | | Authors: | Conti, E, Uy, M, Leighton, L, Blobel, G, Kuriyan, J. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha.
Cell(Cambridge,Mass.), 94, 1998
|
|
6AXF
 
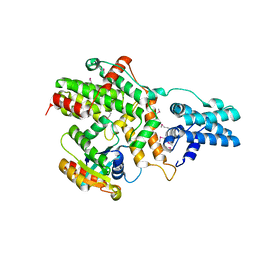 | | Structure of RasGRP2 in complex with Rap1B | | Descriptor: | RAS guanyl-releasing protein 2, Ras-related protein Rap-1b | | Authors: | Kondo, Y, Iwig, J.S, Kuriyan, J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A histidine pH sensor regulates activation of the Ras-specific guanine nucleotide exchange factor RasGRP1.
Elife, 6, 2017
|
|
6AX7
 
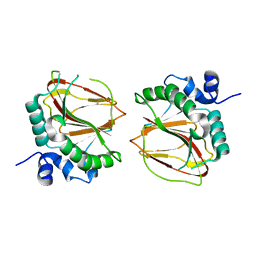 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | Descriptor: | FE (II) ION, Procollagen lysyl hydroxylase and glycosyltransferase | | Authors: | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
2RFD
 
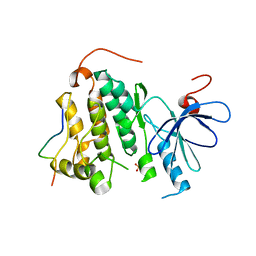 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor, SULFATE ION | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
1PLQ
 
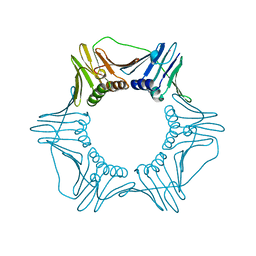 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | MERCURY (II) ION, PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
1PLR
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
6AXG
 
 | | Structure of RasGRP4 in complex with HRas | | Descriptor: | GTPase HRas, RAS guanyl-releasing protein 4 | | Authors: | Kondo, Y, Iwig, J.S, Kuriyan, J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | A histidine pH sensor regulates activation of the Ras-specific guanine nucleotide exchange factor RasGRP1.
Elife, 6, 2017
|
|
6AX6
 
 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | Descriptor: | FE (II) ION, IODIDE ION, Procollagen lysyl hydroxylase and glycosyltransferase | | Authors: | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
1OT3
 
 | | Crystal structure of Drosophila deoxyribonucleotide kinase complexed with the substrate deoxythymidine | | Descriptor: | Deoxyribonucleoside Kinase, SULFATE ION, THYMIDINE | | Authors: | Mikkelsen, N.E, Johansson, K, Karlsson, A, Knecht, W, Andersen, G, Piskur, J, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2003-03-21 | | Release date: | 2003-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Feedback Inhibition of the Deoxyribonucleoside Salvage Pathway: Studies of the Drosophila Deoxyribonucleoside Kinase.
Biochemistry, 42, 2003
|
|
2F4J
 
 | | Structure of the Kinase Domain of an Imatinib-Resistant Abl Mutant in Complex with the Aurora Kinase Inhibitor VX-680 | | Descriptor: | CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Young, M.A, Shah, N.P, Chao, L.H, Zarrinkar, P, Sawyers, P, Kuriyan, J. | | Deposit date: | 2005-11-23 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of the kinase domain of an imatinib-resistant Abl mutant in complex with the Aurora kinase inhibitor VX-680.
Cancer Res., 66, 2006
|
|
1NJF
 
 | | Nucleotide bound form of an isolated E. coli clamp loader gamma subunit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase III subunit gamma, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nucleotide-Induced Conformational Changes in an Isolated Escherichia coli DNA Polymerase III Clamp Loader Subunit
Structure, 11, 2003
|
|
1M52
 
 | | Crystal Structure of the c-Abl Kinase domain in complex with PD173955 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2,6-DICHLORO-PHENYL)-8-METHYL-2-(3-METHYLSULFANYL-PHENYLAMINO)-8H-PYRIDO[2,3-D]PYRIMIDIN-7-ONE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL1 | | Authors: | Nagar, B, Bornmann, W, Pellicena, P, Schindler, T, Veach, D, Miller, W.T, Clarkson, B, Kuriyan, J. | | Deposit date: | 2002-07-08 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Kinase Domain of c-Abl in Complex with the Small Molecule Inhibitors PD173955 and Imatinib (STI-571)
Cancer Res., 62, 2002
|
|
1NVX
 
 | | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Margarit, S.M, Sondermann, H, Hall, B.E, Nagar, B, Hoelz, A, Pirruccello, M, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS
Cell(Cambridge,Mass.), 112, 2003
|
|
1NVW
 
 | | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Margarit, S.M, Sondermann, H, Hall, B.E, Nagar, B, Hoelz, A, Pirruccello, M, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS
Cell(Cambridge,Mass.), 112, 2003
|
|
1NJG
 
 | | Nucleotide-free form of an Isolated E. coli Clamp Loader Gamma Subunit | | Descriptor: | DNA polymerase III subunit gamma, SULFATE ION, ZINC ION | | Authors: | Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nucleotide-Induced Conformational Changes in an Isolated Escherichia coli DNA Polymerase III Clamp Loader Subunit
Structure, 11, 2003
|
|
1OE0
 
 | | CRYSTAL STRUCTURE OF DROSOPHILA DEOXYRIBONUCLEOSIDE KINASE IN COMPLEX WITH DTTP | | Descriptor: | DEOXYRIBONUCLEOSIDE KINASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Mikkelsen, N.E, Johansson, K, Karlsson, A, Knecht, W, Andersen, G, Piskur, J, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2003-03-17 | | Release date: | 2003-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Feedback Inhibition of the Deoxyribonucleoside Salvage Pathway:Studies of the Drosophila Deoxyribonucleoside Kinase
Biochemistry, 42, 2003
|
|
3D1E
 
 | | Crystal structure of E. coli sliding clamp (beta) bound to a polymerase II peptide | | Descriptor: | DNA polymerase III subunit beta, decamer from polymerase II C-terminal | | Authors: | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
