6T6A
 
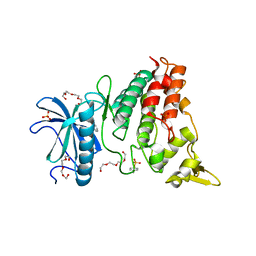 | | Crystal structure of DYRK1A complexed with KuFal319 (compound 11) | | Descriptor: | 4-chloranyl-5~{H}-cyclohepta[b]indol-10-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | [ b ]-Annulated Halogen-Substituted Indoles as Potential DYRK1A Inhibitors.
Molecules, 24, 2019
|
|
9F92
 
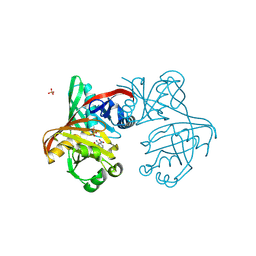 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-3-nitrobenzoic acid | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-azanyl-3-nitro-benzoic acid, SULFATE ION, ... | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
9F96
 
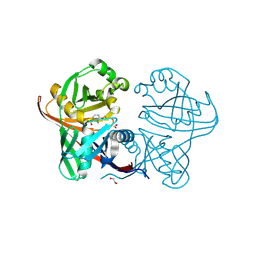 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-3-ethoxybenzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-3-ethoxy-benzoic acid, Trans-2,3-dihydro-3-hydroxyanthranilate isomerase | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
9F93
 
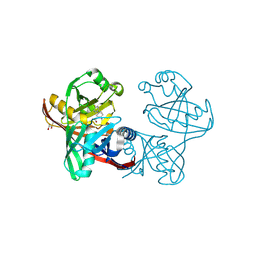 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-5-(4-fluorophenyl)benzoic acid | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-azanyl-5-(4-fluorophenyl)benzoic acid, Trans-2,3-dihydro-3-hydroxyanthranilate isomerase | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
9F94
 
 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-5-(3-hydroxyphenyl)benzoic acid | | Descriptor: | 2-azanyl-5-(3-hydroxyphenyl)benzoic acid, ACETATE ION, FORMIC ACID, ... | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
9F95
 
 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-3-hydroxy-5-(3-hydroxyphenyl)benzoic acid | | Descriptor: | 2-azanyl-5-(3-hydroxyphenyl)-3-oxidanyl-benzoic acid, Trans-2,3-dihydro-3-hydroxyanthranilate isomerase | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
7AYI
 
 | | Crystal structure of Aurora A in complex with 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-one derivative (compound 2a) | | Descriptor: | 7-(2-phenylazanylpyrimidin-4-yl)-1,3,4,5-tetrahydro-1-benzazepin-2-one, Aurora kinase A | | Authors: | Chaikuad, A, Karatas, M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-ones Designed by a "Cut and Glue" Strategy Are Dual Aurora A/VEGF-R Kinase Inhibitors.
Molecules, 26, 2021
|
|
7AYH
 
 | | Crystal structure of Aurora A in complex with 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-one derivative (compound 2c) | | Descriptor: | 7-[2-[(4-methoxyphenyl)amino]pyrimidin-4-yl]-1,3,4,5-tetrahydro-1-benzazepin-2-one, Aurora kinase A | | Authors: | Chaikuad, A, Karatas, M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-ones Designed by a "Cut and Glue" Strategy Are Dual Aurora A/VEGF-R Kinase Inhibitors.
Molecules, 26, 2021
|
|
6FT8
 
 | | Crystal structure of CLK1 in complex with inhibitor 8g | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-hydroxyphenyl)-1~{H}-pyrrolo[3,4-g]indol-8-one, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
6FT9
 
 | | Crystal structure of CLK1 in complex with inhibitor 16 | | Descriptor: | 2-bromanyl-3-phenyl-1~{H}-pyrrolo[3,4-g]indol-8-one, BROMIDE ION, Dual specificity protein kinase CLK1, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
6FT7
 
 | | Crystal structure of CLK3 in complex with compound 8a | | Descriptor: | 1,2-ETHANEDIOL, 3-phenyl-1~{H}-pyrrolo[3,4-g]indol-8-one, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
4YLL
 
 | | Crystal structure of DYRK1AA in complex with 10-Bromo-substituted 11H-indolo[3,2-c]quinolone-6-carboxylic acid inhibitor 5t | | Descriptor: | 1,2-ETHANEDIOL, 10-bromo-2-iodo-11H-indolo[3,2-c]quinoline-6-carboxylic acid, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ... | | Authors: | Chaikuad, A, Falke, H, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-05 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 10-Iodo-11H-indolo[3,2-c]quinoline-6-carboxylic Acids Are Selective Inhibitors of DYRK1A.
J.Med.Chem., 58, 2015
|
|
4YLK
 
 | | Crystal structure of DYRK1A in complex with 10-Chloro-substituted 11H-indolo[3,2-c]quinolone-6-carboxylic acid inhibitor 5s | | Descriptor: | 1,2-ETHANEDIOL, 10-chloro-2-iodo-11H-indolo[3,2-c]quinoline-6-carboxylic acid, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ... | | Authors: | Chaikuad, A, Falke, H, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-05 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 10-Iodo-11H-indolo[3,2-c]quinoline-6-carboxylic Acids Are Selective Inhibitors of DYRK1A.
J.Med.Chem., 58, 2015
|
|
4YLJ
 
 | | Crystal structure of DYRK1A in complex with 10-Iodo-substituted 11H-indolo[3,2-c]quinoline-6-carboxylic acid inhibitor 5j | | Descriptor: | 10-iodo-11H-indolo[3,2-c]quinoline-6-carboxylic acid, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Chaikuad, A, Falke, H, Nowak, R, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-05 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | 10-Iodo-11H-indolo[3,2-c]quinoline-6-carboxylic Acids Are Selective Inhibitors of DYRK1A.
J.Med.Chem., 58, 2015
|
|
