6IDN
 
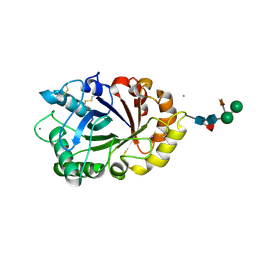 | | Crystal structure of ICChI chitinase from ipomoea carnea | | Descriptor: | CALCIUM ION, ICChI, a glycosylated chitinase, ... | | Authors: | Kumar, S, Kumar, A, Patel, A.K. | | Deposit date: | 2018-09-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | TIM barrel fold and glycan moieties in the structure of ICChI, a protein with chitinase and lysozyme activity.
Phytochemistry, 170, 2020
|
|
3P1B
 
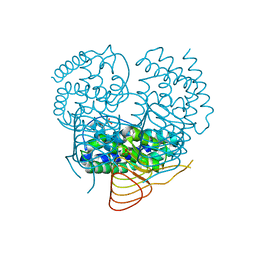 | |
5XYH
 
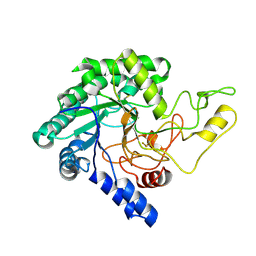 | | Crystal Structure of catalytic domain of 1,4-beta-Cellobiosidase (CbsA) from Xanthomonas oryzae pv. oryzae | | Descriptor: | CbsA | | Authors: | Kumar, S, Haque, A.S, Nathawat, R, Sankaranaryanan, R. | | Deposit date: | 2017-07-07 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | A mutation in an exoglucanase of Xanthomonas oryzae pv. oryzae, which confers an endo mode of activity, affects bacterial virulence, but not the induction of immune responses, in rice
Mol. Plant Pathol., 19, 2018
|
|
3LI6
 
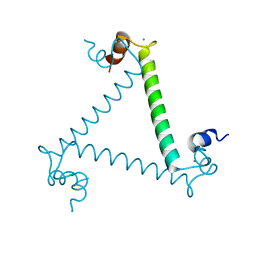 | | Crystal structure and trimer-monomer transition of N-terminal domain of EhCaBP1 from Entamoeba histolytica | | Descriptor: | CALCIUM ION, Calcium-binding protein | | Authors: | Kumar, S, Ahmad, E, Kumar, S, Mansuri, M.S, Khan, R.H, Samudrala, G. | | Deposit date: | 2010-01-24 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structure and trimer-monomer transition of N-terminal domain of EhCaBP1 from Entamoeba histolytica
Biophys.J., 98, 2010
|
|
6UUM
 
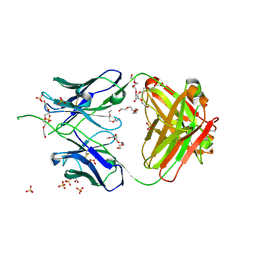 | | Crystal structure of antibody 438-B11 DSS mutant (Cys98A-Cys100aA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, B11 DSS Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
6UUL
 
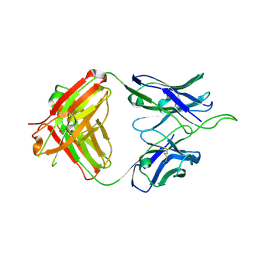 | | Crystal structure of broad and potent HIV-1 neutralizing antibody 438-D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, D5 Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
6UTK
 
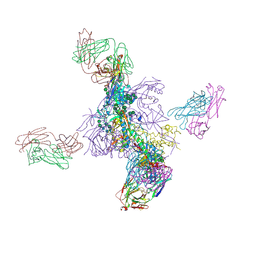 | |
6UUH
 
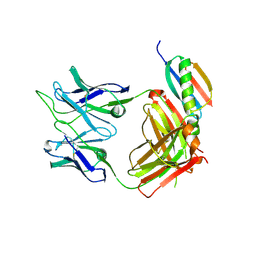 | |
3OIW
 
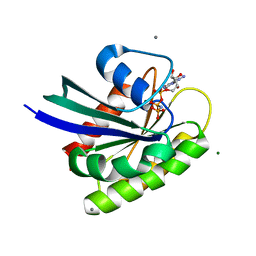 | | H-RasG12V with allosteric switch in the "on" state | | Descriptor: | ACETATE ION, CALCIUM ION, GTPase HRas, ... | | Authors: | Kumar, S, Buhrman, G, Mattos, C. | | Deposit date: | 2010-08-20 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Allosteric Modulation of Ras-GTP Is Linked to Signal Transduction through RAF Kinase.
J.Biol.Chem., 286, 2011
|
|
3PX1
 
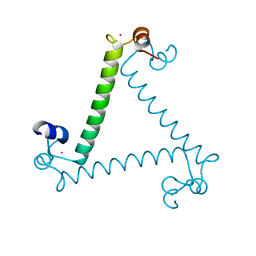 | | Structure of Calcium Binding Protein-1 from Entamoeba histolytica in complex with Strontium | | Descriptor: | Calcium-binding protein, STRONTIUM ION | | Authors: | Kumar, S, Kumar, S, Ahmad, E, Khan, R.H, Gourinath, S. | | Deposit date: | 2010-12-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Flexibility and plasticity of EF-hand motifs: Structure of Calcium Binding Protein-1 from Entamoeba histolytica in complex with Pb2+, Ba2+, and Sr2+.
To be Published
|
|
4H3O
 
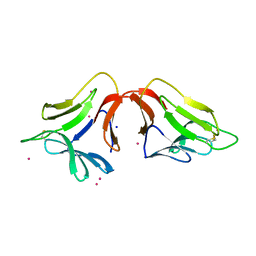 | | Crystal structure of a new form of lectin from Allium sativum at 2.17 A resolution | | Descriptor: | CADMIUM ION, Lectin, SODIUM ION | | Authors: | Kumar, S, Yamini, S, Kumar, J, Kaur, P, Singh, T.P, Dey, S. | | Deposit date: | 2012-09-14 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of a new form of lectin from Allium sativum at 2.17 A resolution
To be Published
|
|
4Y2F
 
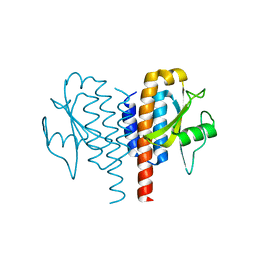 | |
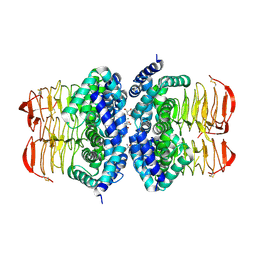
4HZC
 
 | |
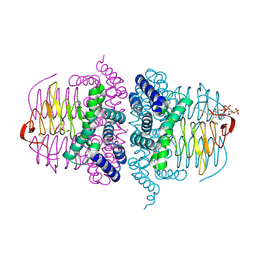
4HZD
 
 | |
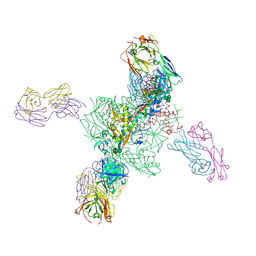
6V6W
 
 | |
6U8T
 
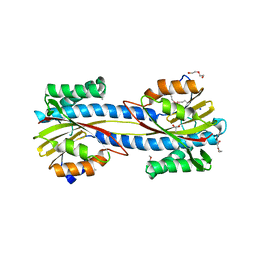 | |
3ULG
 
 | |
1AUL
 
 | | SOLUTION STRUCTURE OF A HIGHLY STABLE DNA DUPLEX CONJUGATED TO A MINOR GROOVE BINDER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*TP*AP*AP*TP*CP*A)-3'), DNA (5'-D(P*THXP*GP*AP*TP*TP*AP*TP*CP*TP*G)-3') | | Authors: | Kumar, S, Reed, M.W, Gamper Junior, H.B, Gorn, V.V, Lukhtanov, E.A, Foti, M, West, J, Meyer Junior, R.B, Schweitzer, B.I. | | Deposit date: | 1997-08-29 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a highly stable DNA duplex conjugated to a minor groove binder.
Nucleic Acids Res., 26, 1998
|
|
3MU7
 
 | | Crystal structure of the xylanase and alpha-amylase inhibitor protein (XAIP-II) from scadoxus multiflorus at 1.2 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, xylanase and alpha-amylase inhibitor protein | | Authors: | Kumar, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-02 | | Release date: | 2010-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Modulation of inhibitory activity of xylanase-alpha-amylase inhibitor protein (XAIP): binding studies and crystal structure determination of XAIP-II from Scadoxus multiflorus at 1.2 A resolution.
Bmc Struct.Biol., 10, 2010
|
|
1F9B
 
 | | MELANIN PROTEIN INTERACTION: X-RAY STRUCTURE OF THE COMPLEX OF MARE LACTOFERRIN WITH MELANIN MONOMERS | | Descriptor: | 3H-INDOLE-5,6-DIOL, BICARBONATE ION, FE (III) ION, ... | | Authors: | Kumar, S, Singh, T.P, Sharma, A.K, Singh, N, Raman, G. | | Deposit date: | 2000-07-10 | | Release date: | 2001-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lactoferrin-melanin interaction and its possible implications in melanin polymerization: crystal structure of the complex formed between mare lactoferrin and melanin monomers at 2.7-A resolution.
Proteins, 45, 2001
|
|
6CE0
 
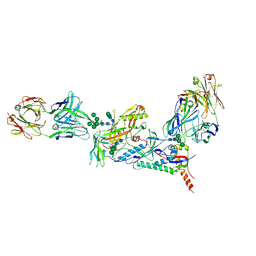 | | Crystal structure of a HIV-1 clade B tier-3 isolate H078.14 UFO-BG Env trimer in complex with broadly neutralizing Fabs PGT124 and 35O22 at 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-05 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (4.602 Å) | | Cite: | HIV-1 vaccine design through minimizing envelope metastability.
Sci Adv, 4, 2018
|
|
3M7S
 
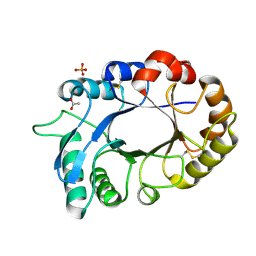 | | Crystal structure of the complex of xylanase GH-11 and alpha amylase inhibitor protein with cellobiose at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, S, Dube, D, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
6CA8
 
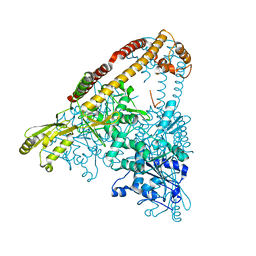 | |
7MK2
 
 | | CryoEM Structure of NPR1 | | Descriptor: | Regulatory protein NPR1, ZINC ION | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M, Bartesaghi, A, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
8DTI
 
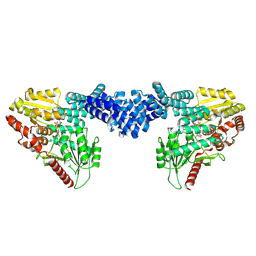 | | Cryo-EM structure of Arabidopsis SPY in complex with GDP-fucose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
