1AC9
 
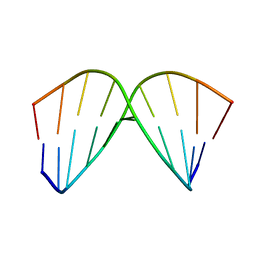 | | SOLUTION STRUCTURE OF A DNA DECAMER CONTAINING THE ANTIVIRAL DRUG GANCICLOVIR: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS, AND FULL RELAXATION REFINEMENT, 6 STRUCTURES | | Descriptor: | DNA | | Authors: | Foti, M, Marshalko, S, Schurter, E, Kumar, S, Beardsley, G.P, Schweitzer, B.I. | | Deposit date: | 1997-02-17 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA decamer containing the antiviral drug ganciclovir: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 36, 1997
|
|
3NOE
 
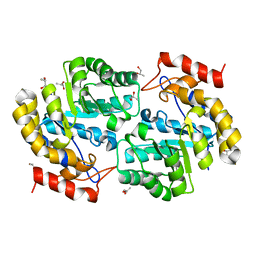 | | Crystal Structure of Dihydrodipicolinate synthase from Pseudomonas aeruginosa | | Descriptor: | Dihydrodipicolinate synthase, S-1,2-PROPANEDIOL | | Authors: | Kaur, N, Kumar, S, Singh, N, Gautam, A, Sharma, R, Sharma, S, Tewari, R, Singh, T.P. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate synthase from Pseudomonas aeruginosa
To be Published
|
|
1DQ7
 
 | | THREE-DIMENSIONAL STRUCTURE OF A NEUROTOXIN FROM RED SCORPION (BUTHUS TAMULUS) AT 2.2A RESOLUTION. | | Descriptor: | NEUROTOXIN | | Authors: | Sharma, M, Yadav, S, Karthikeyan, S, Kumar, S, Paramasivam, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 1999-12-30 | | Release date: | 2000-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional Structure of a Neurotoxin from Red Scorpion (Buthus tamulus) at 2.2A Resolution
To be Published
|
|
3TW7
 
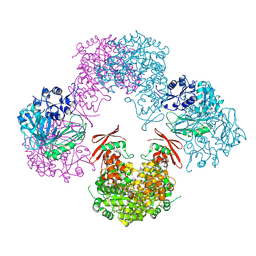 | | Structure of Rhizobium etli pyruvate carboxylase T882A crystallized without acetyl coenzyme-A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Pyruvate carboxylase protein, ... | | Authors: | St Maurice, M, Kumar, S, Lietzan, A.D. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-12 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli.
Biochemistry, 50, 2011
|
|
8TQS
 
 | |
2LXN
 
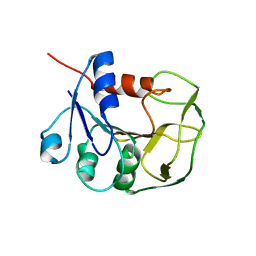 | | Solution NMR structure of glutamine amido transferase subunit of gaunosine monophosphate synthetase from Methanocaldococcus jannaschii | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit A | | Authors: | Ali, R, Kumar, S, Balaram, H, Sarma, S.P. | | Deposit date: | 2012-08-30 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C, 15N assignment and secondary structure determination of glutamine amido transferase subunit of gaunosine monophosphate synthetase from Methanocaldococcus jannaschii
Biomol.Nmr Assign., 6, 2012
|
|
8XEQ
 
 | |
8XGW
 
 | |
1RGZ
 
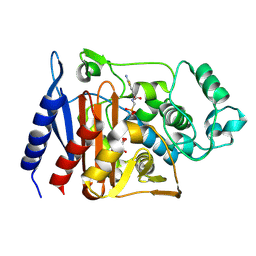 | | Enterobacter cloacae GC1 Class C beta-Lactamase Complexed with Transition-State Analog of Cefotaxime | | Descriptor: | GLYCEROL, class C beta-lactamase, {[(2E)-2-(2-AMINO-1,3-THIAZOL-4-YL)-2-(METHOXYIMINO)ETHANOYL]AMINO}METHYLPHOSPHONIC ACID | | Authors: | Nukaga, M, Kumar, S, Nukaga, K, Pratt, R.F, Knox, J.R. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Hydrolysis of third-generation cephalosporins by class C beta-lactamases. Structures of a transition state analog of cefotoxamine in wild-type and extended spectrum enzymes.
J.Biol.Chem., 279, 2004
|
|
2K8M
 
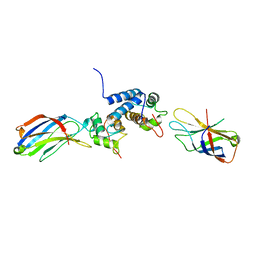 | | S100A13-C2A binary complex structure | | Descriptor: | Protein S100-A13, Putative uncharacterized protein | | Authors: | Mohan, S.K, Rani, S.G, Kumar, S.M, Yu, C. | | Deposit date: | 2008-09-14 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | S100A13-C2A binary complex structure-a key component in the acidic fibroblast growth factor for the non-classical pathway.
Biochem.Biophys.Res.Commun., 380, 2009
|
|
3PH0
 
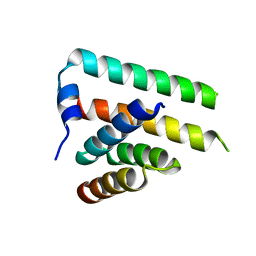 | | Crystal structure of the heteromolecular chaperone, AscE-AscG, from the type III secretion system in Aeromonas hydrophila | | Descriptor: | AscE, AscG | | Authors: | Chatterjee, C, Kumar, S, Chakraborty, S, Tan, Y.W, Leung, K.Y, Sivaraman, J, Mok, Y.K. | | Deposit date: | 2010-11-03 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the heteromolecular chaperone, AscE-AscG, from the type III secretion system in Aeromonas hydrophila
Plos One, 6, 2011
|
|
7CCA
 
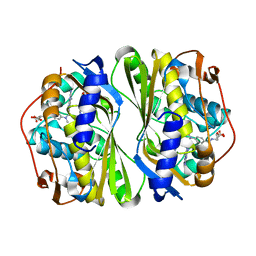 | | Crystal structure of White Spot Syndrome Virus Thymidylate Synthase - ternary complex with Methotrexate and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, METHOTREXATE, Thymidylate Synthase | | Authors: | Panchal, N.V, Kumar, S, Shaikh, N, Vasudevan, D. | | Deposit date: | 2020-06-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure analysis of thymidylate synthase from white spot syndrome virus reveals WSSV-specific structural elements.
Int.J.Biol.Macromol., 167, 2021
|
|
3H8B
 
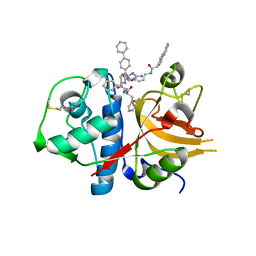 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors(compound 9) | | Descriptor: | Cathepsin L1, N~2~,N~6~-bis(biphenyl-4-ylacetyl)-L-lysyl-D-arginyl-N-(2-phenylethyl)-L-phenylalaninamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A combined crystallographic and molecular dynamics study of cathepsin L retrobinding inhibitors
J.Med.Chem., 52, 2009
|
|
3H89
 
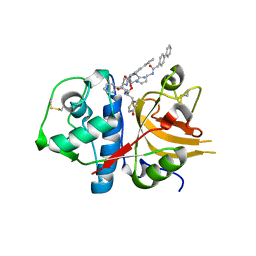 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors(compound 4) | | Descriptor: | Cathepsin L1, N~2~,N~6~-bis(biphenyl-4-ylacetyl)-L-lysyl-D-arginyl-N-(2-phenylethyl)-L-tyrosinamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A combined crystallographic and molecular dynamics study of cathepsin L retrobinding inhibitors
J.Med.Chem., 52, 2009
|
|
4FYM
 
 | |
4N42
 
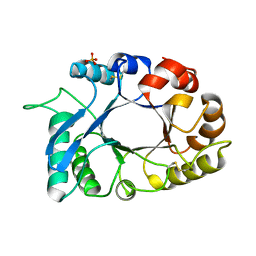 | | Crystal structure of allergen protein scam1 from Scadoxus multiflorus | | Descriptor: | PHOSPHATE ION, Xylanase and alpha-amylase inhibitor protein isoform III | | Authors: | Singh, A, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of allergen protein scam1 from Scadoxus multiflorus
To be published
|
|
7EWO
 
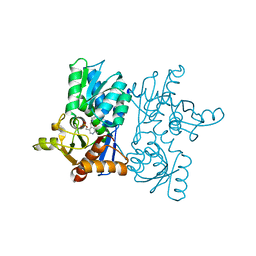 | | Crystal Structure of D67A, E68P double mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae | | Descriptor: | Cysteine synthase | | Authors: | Rahisuddin, R, Ekka, M.K, Singh, A.K, Saini, N, Patel, M, Kumar, N, Kumaran, S. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of D67A, E68P double mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae
To Be Published
|
|
3TW6
 
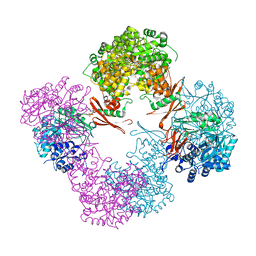 | | Structure of Rhizobium etli pyruvate carboxylase T882A with the allosteric activator, acetyl coenzyme-A | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | St Maurice, M, Kumar, S, Lietzan, A.D. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-19 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli.
Biochemistry, 50, 2011
|
|
7D40
 
 | |
4NZC
 
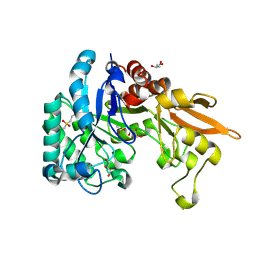 | | Crystal structure of Chitinase D from Serratia proteamaculans at 1.45 Angstrom resolution | | Descriptor: | ACETATE ION, GLYCEROL, Glycoside hydrolase family 18 | | Authors: | Madhuprakash, J, Singh, A, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Podile, A.R, Singh, T.P. | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of chitinase D from Serratia proteamaculans reveals the structural basis of its dual action of hydrolysis and transglycosylation
Int J Biochem Mol Biol, 4, 2013
|
|
3OQT
 
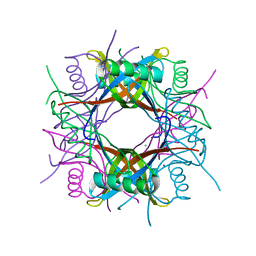 | | Crystal structure of Rv1498A protein from mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, Rv1498A PROTEIN, SODIUM ION | | Authors: | Liu, F, Xiong, J, Kumar, S, Yang, C, Li, S, Ge, S, Xia, N, Swaminathan, K. | | Deposit date: | 2010-09-04 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural and biophysical characterization of Mycobacterium tuberculosis dodecin Rv1498A.
J.Struct.Biol., 175, 2011
|
|
3K1D
 
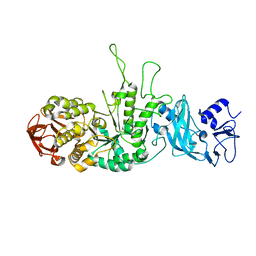 | | Crystal structure of glycogen branching enzyme synonym: 1,4-alpha-D-glucan:1,4-alpha-D-GLUCAN 6-glucosyl-transferase from mycobacterium tuberculosis H37RV | | Descriptor: | 1,4-alpha-glucan-branching enzyme | | Authors: | Pal, K, Kumar, S, Swaminathan, K. | | Deposit date: | 2009-09-27 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of full-length Mycobacterium tuberculosis H37Rv glycogen branching enzyme: insights of N-terminal beta-sandwich in substrate specificity and enzymatic activity
J.Biol.Chem., 285, 2010
|
|
3E9K
 
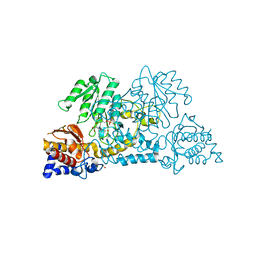 | | Crystal structure of Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex | | Descriptor: | 3-Hydroxyhippuric acid, Kynureninase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lima, S, Kumar, S, Gawandi, V, Momany, C, Phillips, R.S. | | Deposit date: | 2008-08-22 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex: insights into the molecular basis of kynureninase substrate specificity.
J.Med.Chem., 52, 2009
|
|
2FHT
 
 | | Crystal Structure of Viral Macrophage Inflammatory Protein-II | | Descriptor: | Viral macrophage inflammatory protein-II | | Authors: | Li, Y, Liu, D, Cao, R, Kumar, S, Dong, C.Z, wilson, S.R, Gao, Y.G, Huang, Z. | | Deposit date: | 2005-12-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chemically synthesized vMIP-II.
Proteins, 67, 2007
|
|
7D95
 
 | |
