3D5H
 
 | | Crystal structure of haementhin from Haemanthus multiflorus at 2.0A resolution: Formation of a novel loop on a TIM barrel fold and its functional significance | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Kumar, S, Singh, N, Sinha, M, Singh, S.B, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2008-05-16 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of haementhin from Haemanthus multiflorus at 2.0A resolution: Formation of a novel loop on a TIM barrel fold and its functional significance
To be Published
|
|
7D5Q
 
 | | Structure of NorC transporter (K398A mutant) in an outward-open conformation in complex with a single-chain Indian camelid antibody | | Descriptor: | Drug transporter, putative, ICab, ... | | Authors: | Kumar, S, Athreya, A, Penmatsa, A. | | Deposit date: | 2020-09-27 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of inhibition of a transporter from Staphylococcus aureus, NorC, through a single-domain camelid antibody.
Commun Biol, 4, 2021
|
|
7D5P
 
 | | Structure of NorC transporter in an outward-open conformation in complex with a single-chain Indian camelid antibody | | Descriptor: | Drug transporter, putative, ICab, ... | | Authors: | Kumar, S, Athreya, A, Penmatsa, A. | | Deposit date: | 2020-09-27 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis of inhibition of a transporter from Staphylococcus aureus, NorC, through a single-domain camelid antibody.
Commun Biol, 4, 2021
|
|
3LI6
 
 | | Crystal structure and trimer-monomer transition of N-terminal domain of EhCaBP1 from Entamoeba histolytica | | Descriptor: | CALCIUM ION, Calcium-binding protein | | Authors: | Kumar, S, Ahmad, E, Kumar, S, Mansuri, M.S, Khan, R.H, Samudrala, G. | | Deposit date: | 2010-01-24 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structure and trimer-monomer transition of N-terminal domain of EhCaBP1 from Entamoeba histolytica
Biophys.J., 98, 2010
|
|
3PX1
 
 | | Structure of Calcium Binding Protein-1 from Entamoeba histolytica in complex with Strontium | | Descriptor: | Calcium-binding protein, STRONTIUM ION | | Authors: | Kumar, S, Kumar, S, Ahmad, E, Khan, R.H, Gourinath, S. | | Deposit date: | 2010-12-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Flexibility and plasticity of EF-hand motifs: Structure of Calcium Binding Protein-1 from Entamoeba histolytica in complex with Pb2+, Ba2+, and Sr2+.
To be Published
|
|
4H3O
 
 | | Crystal structure of a new form of lectin from Allium sativum at 2.17 A resolution | | Descriptor: | CADMIUM ION, Lectin, SODIUM ION | | Authors: | Kumar, S, Yamini, S, Kumar, J, Kaur, P, Singh, T.P, Dey, S. | | Deposit date: | 2012-09-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of a new form of lectin from Allium sativum at 2.17 A resolution
To be Published
|
|
1F9B
 
 | | MELANIN PROTEIN INTERACTION: X-RAY STRUCTURE OF THE COMPLEX OF MARE LACTOFERRIN WITH MELANIN MONOMERS | | Descriptor: | 3H-INDOLE-5,6-DIOL, BICARBONATE ION, FE (III) ION, ... | | Authors: | Kumar, S, Singh, T.P, Sharma, A.K, Singh, N, Raman, G. | | Deposit date: | 2000-07-10 | | Release date: | 2001-02-10 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lactoferrin-melanin interaction and its possible implications in melanin polymerization: crystal structure of the complex formed between mare lactoferrin and melanin monomers at 2.7-A resolution.
Proteins, 45, 2001
|
|
4YEH
 
 | | Crystal structure of Mg2+ ion containing hemopexin fold from Kabuli chana (chickpea white) at 2.45A resolution reveals a structural basis of metal ion transport | | Descriptor: | Lectin, MAGNESIUM ION | | Authors: | Kumar, S, Singh, A, Yamini, S, Bhushan, A, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Mg(2+) Containing Hemopexin-Fold Protein from Kabuli Chana (Chickpea-White, CW-25) at 2.45 angstrom Resolution Reveals Its Metal Ion Transport Property
Protein J., 34, 2015
|
|
4YDO
 
 | | CRYSTAL STRUCTURE OF CANDIDA ALBICANS PROTEIN FARNESYLTRANSFERASE IN APO FORM | | Descriptor: | CALCIUM ION, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, Uncharacterized protein, ... | | Authors: | Kumar, S, Mabanglo, M.F, Hast, M.A, Shi, Y, Beese, L.S. | | Deposit date: | 2015-02-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | CRYSTAL STRUCTURE OF CANDIDA ALBICANS PROTEIN FARNESYLTRANSFERASE IN APO FORM
To Be Published
|
|
6U8T
 
 | |
4YDE
 
 | | CRYSTAL STRUCTURE OF CANDIDA ALBICANS PROTEIN FARNESYLTRANSFERASE BINARY COMPLEX WITH THE ISOPRENOID FARNESYLDIPHOSPHATE | | Descriptor: | (3R,7S)-3,7,11-trimethyldodecyl trihydrogen diphosphate, 1,2-ETHANEDIOL, Protein farnesyltransferase/geranylgeranyltransferase type-1 Subunit beta, ... | | Authors: | Kumar, S, Mabanglo, M.F, Hast, M.A, Shi, Y, Beese, L.S. | | Deposit date: | 2015-02-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | CRYSTAL STRUCTURE OF CANDIDA ALBICANS PROTEIN FARNESYLTRANSFERASE BINARY COMPLEX WITH THE ISOPRENOID FARNESYLDIPHOSPHATE
To Be Published
|
|
7CC8
 
 | | Crystal structure of White Spot Syndrome Virus Thymidylate Synthase - Apo form | | Descriptor: | SULFATE ION, TRIETHYLENE GLYCOL, Thymidylate Synthase | | Authors: | Kumar, S, Panchal, N.V, Shaikh, N, Vasudevan, D. | | Deposit date: | 2020-06-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure analysis of thymidylate synthase from white spot syndrome virus reveals WSSV-specific structural elements.
Int.J.Biol.Macromol., 167, 2021
|
|
7CB8
 
 | | Structure of a FIC-domain protein from Mycobacterium marinum in complex with CDP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, S, Singh, A, Penmatsa, A, Surolia, A. | | Deposit date: | 2020-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of of a FIC-domain protein from Mycobacterium marinum
To Be Published
|
|
4HZC
 
 | |
4HZD
 
 | |
4Y2F
 
 | |
6UUM
 
 | | Crystal structure of antibody 438-B11 DSS mutant (Cys98A-Cys100aA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, B11 DSS Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
3HU7
 
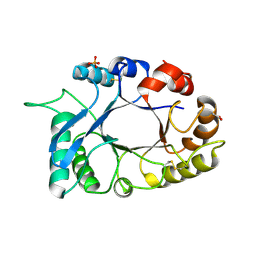 | | Structural characterization and binding studies of a plant pathogenesis related protein heamanthin from haemanthus multiflorus reveal its dual inhibitory effects against xylanase and alpha-amylase | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Kumar, S, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-06-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
6MCO
 
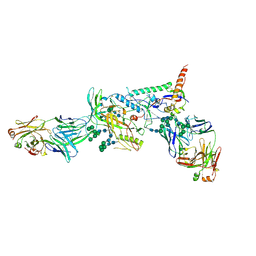 | | Crystal structure of the B41 SOSIP.664 Env trimer with PGT124 and 35O22 Fabs, in P23 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-08-31 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Capturing the inherent structural dynamics of the HIV-1 envelope glycoprotein fusion peptide.
Nat Commun, 10, 2019
|
|
6UTK
 
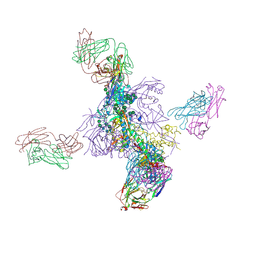 | |
6MDT
 
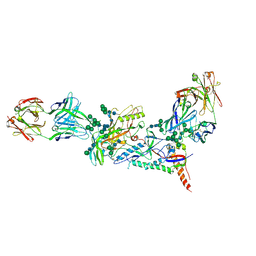 | | Crystal structure of the B41 SOSIP.664 Env trimer with PGT124 and 35O22 Fabs, in P63 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.816 Å) | | Cite: | Capturing the inherent structural dynamics of the HIV-1 envelope glycoprotein fusion peptide.
Nat Commun, 10, 2019
|
|
6V6W
 
 | |
3OIW
 
 | | H-RasG12V with allosteric switch in the "on" state | | Descriptor: | ACETATE ION, CALCIUM ION, GTPase HRas, ... | | Authors: | Kumar, S, Buhrman, G, Mattos, C. | | Deposit date: | 2010-08-20 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Allosteric Modulation of Ras-GTP Is Linked to Signal Transduction through RAF Kinase.
J.Biol.Chem., 286, 2011
|
|
6ME1
 
 | |
6UUL
 
 | | Crystal structure of broad and potent HIV-1 neutralizing antibody 438-D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, D5 Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
