1QYS
 
 | | Crystal structure of Top7: A computationally designed protein with a novel fold | | Descriptor: | TOP7 | | Authors: | Kuhlman, B, Dantas, G, Ireton, G.C, Varani, G, Stoddard, B.L, Baker, D. | | Deposit date: | 2003-09-11 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of a Novel Globular Protein Fold with Atomic-Level Accuracy
Science, 302, 2003
|
|
3ZY7
 
 | | Crystal structure of computationally redesigned gamma-adaptin appendage domain forming a symmetric homodimer | | Descriptor: | AP-1 COMPLEX SUBUNIT GAMMA-1, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL | | Authors: | Stranges, P.B, Machius, M, Miley, M.J, Tripathy, A, Kuhlman, B. | | Deposit date: | 2011-08-17 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Computational Design of a Symmetric Homodimer Using Beta-Strand Assembly.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4WF0
 
 | |
8VY4
 
 | | Engineering a Tumor-Selective Prodrug T Cell Engager Bispecific Antibody for Safer Immunotherapy | | Descriptor: | Fab Heavy Chain, Fab Light Chain, PCA-ASP-GLY-ASN-GLU-GLU-MET | | Authors: | Antonysamy, S, Demarest, S, Froning, K, Hickey, M.H, Kuhlman, B, McCue, A.C. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering a tumor-selective prodrug T-cell engager bispecific antibody for safer immunotherapy.
Mabs, 16, 2024
|
|
6WY1
 
 | | Crystal structure of an engineered thermostable dengue virus 2 envelope protein dimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dengue 2 soluble recombinant envelope | | Authors: | Kudlacek, S.T, Lakshmanane, P, Kuhlman, B. | | Deposit date: | 2020-05-12 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Designed, highly expressing, thermostable dengue virus 2 envelope protein dimers elicit quaternary epitope antibodies.
Sci Adv, 7, 2021
|
|
1CQU
 
 | |
6U07
 
 | | Computational Stabilization of T Cell Receptor Constant Domains | | Descriptor: | MAGNESIUM ION, Stabilized T cell receptor constant domain (Calpha), Stabilized T cell receptor constant domain (Cbeta) | | Authors: | Froning, K, Maguire, J, Sereno, A, Huang, F, Chang, S, Weichert, K, Frommelt, A.J, Dong, J, Wu, X, Austin, H, Conner, E.M, Fitchett, J.R, Heng, A.R, Balasubramaniam, D, Hilgers, M.T, Kuhlman, B, Demarest, S.J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics.
Nat Commun, 11, 2020
|
|
7TJL
 
 | |
3ONW
 
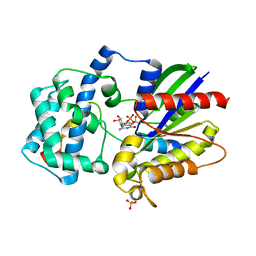 | | Structure of a G-alpha-i1 mutant with enhanced affinity for the RGS14 GoLoco motif. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, Regulator of G-protein signaling 14, ... | | Authors: | Bosch, D, Kimple, A.J, Sammond, D.W, Miley, M.J, Machius, M, Kuhlman, B, Willard, F.S, Siderovski, D.P. | | Deposit date: | 2010-08-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Determinants of Affinity Enhancement between GoLoco Motifs and G-Protein {alpha} Subunit Mutants.
J.Biol.Chem., 286, 2011
|
|
1MI0
 
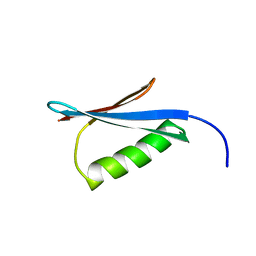 | | Crystal Structure of the redesigned protein G variant NuG2 | | Descriptor: | immunoglobulin-binding protein G | | Authors: | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Biochemistry, 11, 2002
|
|
2XNS
 
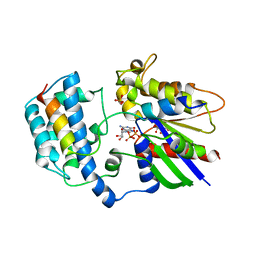 | | Crystal Structure Of Human G alpha i1 Bound To A Designed Helical Peptide Derived From The Goloco Motif Of RGS14 | | Descriptor: | GUANINE NUCLEOTIDE-BINDING PROTEIN G(I) SUBUNIT ALPHA-1, GUANOSINE-5'-DIPHOSPHATE, REGULATOR OF G-PROTEIN SIGNALING 14, ... | | Authors: | Bosch, D, Sammond, D.W, Butterfoss, G.L, Machius, M, Siderovski, D.P, Kuhlman, B. | | Deposit date: | 2010-08-05 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Computational Design of the Sequence and Structure of a Protein-Binding Peptide.
J.Am.Chem.Soc., 133, 2011
|
|
7T2Y
 
 | | X-ray structure of a designed cold unfolding four helix bundle | | Descriptor: | Designed cold unfolding four helix bundle | | Authors: | Harrison, J.S, Kuhlman, B, Szyperski, T, Premkumar, L, Maguire, J, Pulavarti, S, Yuen, S. | | Deposit date: | 2021-12-06 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
3B83
 
 | |
2LCH
 
 | | Solution NMR Structure of a Protein With a Redesigned Hydrophobic Core, Northeast Structural Genomics Consortium Target OR38 | | Descriptor: | Protein OR38 | | Authors: | Mills, J.L, Murphy, G, Miley, M, Machius, M, Kuhlman, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Protein With a Redesigned Hydrophobic Core, Northeast Structural Genomics Consortium Target OR38
To be Published
|
|
7T03
 
 | | NMR structure of a designed cold unfolding four helix bundle | | Descriptor: | Cold unfolding four helix bundle | | Authors: | Pulavarti, S, Szyperski, T, Yuen, S, Maguire, J, Griffin, J, Kuhlman, B. | | Deposit date: | 2021-11-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
5KAY
 
 | | Structure of Spelter bound to Zn2+ | | Descriptor: | SODIUM ION, Spelter, ZINC ION | | Authors: | Guffy, S.L, Der, B.S, Kuhlman, B. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-03 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the minimal determinants of zinc binding with computational protein design.
Protein Eng.Des.Sel., 29, 2016
|
|
5KIZ
 
 | |
1JML
 
 | | Conversion of Monomeric Protein L to an Obligate Dimer by Computational Protein Design | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kuhlman, B, Kim, D.E, Zhang, K.Y.J, Baker, D. | | Deposit date: | 2001-07-19 | | Release date: | 2001-10-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conversion of monomeric protein L to an obligate dimer by computational protein design.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1KH0
 
 | | Accurate Computer Base Design of a New Backbone Conformation in the Second Turn of Protein L | | Descriptor: | protein L | | Authors: | O'Neill, J.W, Kuhlman, B, Kim, D.E, Zhang, K.Y, Baker, D. | | Deposit date: | 2001-11-28 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Accurate computer-based design of a new backbone conformation in the second turn of protein L.
J.Mol.Biol., 315, 2002
|
|
1MHX
 
 | | Crystal Structures of the redesigned protein G variant NuG1 | | Descriptor: | immunoglobulin-binding protein G | | Authors: | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Protein Sci., 11, 2002
|
|
2RB8
 
 | |
2N8I
 
 | | Solution NMR Structure of Designed Protein DA05, Northeast Structural Genomics Consortium (NESG) Target OR626 | | Descriptor: | Designed Protein DA05 | | Authors: | Xu, X, Eletsky, A, Federizon, J.F, Jacobs, T.M, Kuhlman, B, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-10-15 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of structurally distinct proteins using strategies inspired by evolution.
Science, 352, 2016
|
|
2N8W
 
 | | Solution NMR Structure of Designed Protein DA05R1, Northeast Structural Genomics Consortium (NESG) Target OR690 | | Descriptor: | Designed Protein DA05R1 | | Authors: | Eletsky, A, Federizon, J.F, Xu, X, Pulavarti, S, Jacobs, T.M, Kuhlman, B, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of structurally distinct proteins using strategies inspired by evolution.
Science, 352, 2016
|
|
2WKR
 
 | | Structure of a photoactivatable Rac1 containing the Lov2 C450M Mutant | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, Y.I, Frey, D, Lungu, O.I, Jaehrig, A, Schlichting, I, Kuhlman, B, Hahn, K.M. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Genetically Encoded Photoactivatable Rac Controls the Motility of Living Cells.
Nature, 461, 2009
|
|
2WKP
 
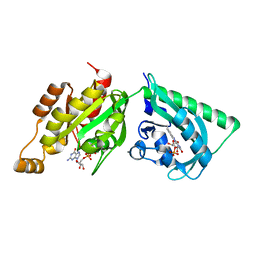 | | Structure of a photoactivatable Rac1 containing Lov2 Wildtype | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, Y.I, Frey, D, Lungu, O.I, Jaehrig, A, Schlichting, I, Kuhlman, B, Hahn, K.M. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Genetically Encoded Photoactivatable Rac Controls the Motility of Living Cells.
Nature, 461, 2009
|
|
