6TWB
 
 | |
6TWC
 
 | |
4INZ
 
 | | The crystal structure of M145A mutant of an epoxide hydrolase from Bacillus megaterium | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Soluble epoxide hydrolase | | Authors: | Kong, X.D, Zhou, J.H, Xu, J.H. | | Deposit date: | 2013-01-07 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4IO0
 
 | | Crystal structure of F128A mutant of an epoxide hydrolase from Bacillus megaterium complexed with its product (R)-3-[1]naphthyloxy-propane-1,2-diol | | Descriptor: | (2R)-3-(naphthalen-1-yloxy)propane-1,2-diol, SULFATE ION, Soluble epoxide hydrolase | | Authors: | Kong, X.D, Zhou, J.H, Xu, J.H. | | Deposit date: | 2013-01-07 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4XBX
 
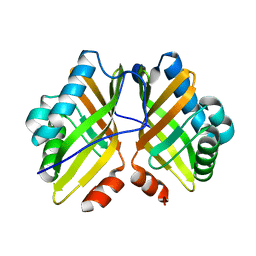 | | Crystal Structure of the L74F/M78F/L103V/L114V/I116V/F139V/L147V mutant of LEH | | Descriptor: | Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XBT
 
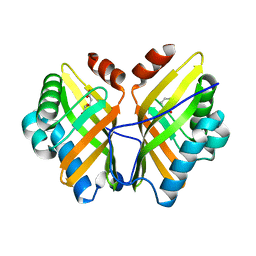 | | Crystal Structure of the L74F/M78F/L103V/L114V/I116V/F139V/L147V mutant of LEH complexed with (S,S)-cyclohexanediol | | Descriptor: | (1S,2S)-cyclohexane-1,2-diol, CITRATE ANION, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XDV
 
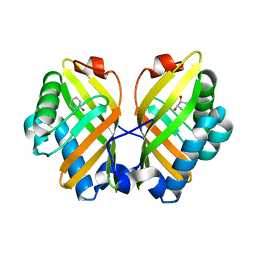 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclohexanediol | | Descriptor: | (1R,2R)-cyclohexane-1,2-diol, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XBY
 
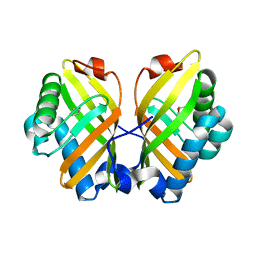 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclopentene oxide | | Descriptor: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XDW
 
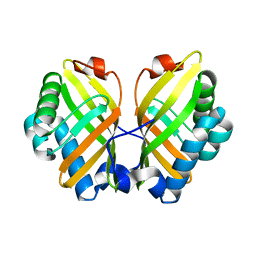 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH | | Descriptor: | Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4NZZ
 
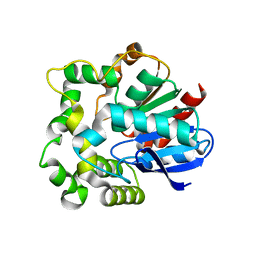 | |
4O08
 
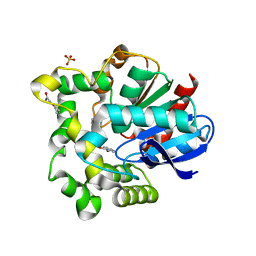 | |
5YQT
 
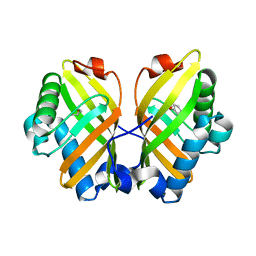 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclopentene oxide | | Descriptor: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z.T, Wu, L, Reetz, M.T, Zhou, J.H, Xu, J.H. | | Deposit date: | 2017-11-07 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Computational Insight into the Catalytic Mechanism of Limonene Epoxide Hydrolase Mutants in Stereoselective Transformations.
J. Am. Chem. Soc., 140, 2018
|
|
5XMD
 
 | |
8WV3
 
 | |
8K56
 
 | |
8K51
 
 | |
4RNC
 
 | | Crystal structure of an esterase RhEst1 from Rhodococcus sp. ECU1013 | | Descriptor: | Esterase, PHOSPHATE ION | | Authors: | Dou, S, Kong, X.D, Xu, J.H, Zhou, J. | | Deposit date: | 2014-10-23 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate channel evolution of an esterase for the synthesis of Cilastatin
CATALYSIS SCIENCE AND TECHNOLOGY, 5, 2015
|
|
8WNH
 
 | |
8WMT
 
 | |
8WWS
 
 | | Crystal structure of cis-epoxysuccinate hydrolase from Klebsiella oxytoca with L(+)-tartaric acid | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Han, Y, Kong, X.D, Li, J, Xu, J.H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Insights of a cis -Epoxysuccinate Hydrolase Facilitate the Development of Robust Biocatalysts for the Production of l-(+)-Tartrate.
Biochemistry, 63, 2024
|
|
4OB7
 
 | | Crystal structure of esterase rPPE mutant W187H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4OB8
 
 | | Crystal structure of a novel thermostable esterase from Pseudomonas putida ECU1011 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4OU5
 
 | | Crystal structure of esterase rPPE mutant S159A/W187H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-02-15 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4O98
 
 | | Crystal structure of Pseudomonas oleovorans PoOPH mutant H250I/I263W | | Descriptor: | ZINC ION, organophosphorus hydrolase | | Authors: | Luo, X.J, Kong, X.D, Zhao, J, Chen, Q, Zhou, J.H, Xu, J.H. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Switching a newly discovered lactonase into an efficient and thermostable phosphotriesterase by simple double mutations His250Ile/Ile263Trp
Biotechnol.Bioeng., 111, 2014
|
|
4OB6
 
 | | Complex structure of esterase rPPE S159A/W187H and substrate (S)-Ac-CPA | | Descriptor: | (2S)-(acetyloxy)(2-chlorophenyl)ethanoic acid, Alpha/beta hydrolase fold-3 domain protein | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Chen, Q, Zhou, J.H, Xu, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
