6UAN
 
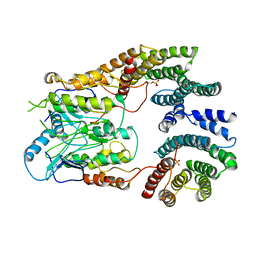 | | B-Raf:14-3-3 complex | | Descriptor: | 14-3-3 zeta, Serine/threonine-protein kinase B-raf | | Authors: | Kondo, Y, Ognjenovic, J, Banerjee, S, Karandur, D, Merk, A, Kulhanek, K, Wong, K, Roose, J.P, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases.
Science, 366, 2019
|
|
5WDR
 
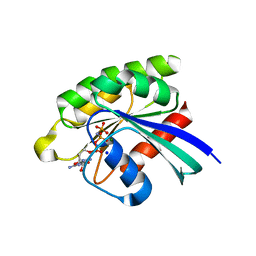 | | Choanoflagellate Salpingoeca rosetta Ras with GMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras protein, ... | | Authors: | Kondo, Y, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
5WDS
 
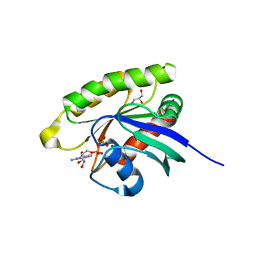 | | Choanoflagellate Salpingoeca rosetta Ras with GDP bound | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kondo, Y, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
6AXF
 
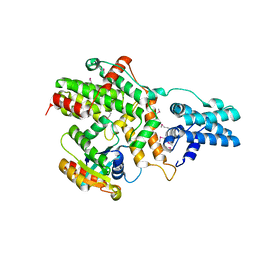 | | Structure of RasGRP2 in complex with Rap1B | | Descriptor: | RAS guanyl-releasing protein 2, Ras-related protein Rap-1b | | Authors: | Kondo, Y, Iwig, J.S, Kuriyan, J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A histidine pH sensor regulates activation of the Ras-specific guanine nucleotide exchange factor RasGRP1.
Elife, 6, 2017
|
|
6AXG
 
 | | Structure of RasGRP4 in complex with HRas | | Descriptor: | GTPase HRas, RAS guanyl-releasing protein 4 | | Authors: | Kondo, Y, Iwig, J.S, Kuriyan, J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | A histidine pH sensor regulates activation of the Ras-specific guanine nucleotide exchange factor RasGRP1.
Elife, 6, 2017
|
|
4PJO
 
 | | Minimal U1 snRNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ETHANOL, ... | | Authors: | Kondo, Y, Oubridge, C, van Roon, A.M, Nagai, K. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5' splice site recognition.
Elife, 4, 2015
|
|
1WRU
 
 | | Structure of central hub elucidated by X-ray analysis of gene product 44; baseplate component of bacteriophage Mu | | Descriptor: | 43 kDa tail protein | | Authors: | Kondou, Y, Kitazawa, D, Takeda, S, Yamashita, E, Mizuguchi, M, Kawano, K, Tsukihara, T. | | Deposit date: | 2004-10-27 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of central hub elucidated by X-ray analysis of gene product 44; baseplate component of bacteriophage Mu
To be Published
|
|
2RT8
 
 | | Structure of metallo-dna in solution | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*CP*GP*CP*G)-3'), MERCURY (II) ION | | Authors: | Yamaguchi, H, Sebera, J, Kondo, J, Oda, S, Komuro, T, Kawamura, T, Dairaku, T, Kondo, Y, Okamoto, I, Ono, A, Burda, J.V, Kojima, C, Sychrovsky, V, Tanaka, Y. | | Deposit date: | 2013-06-18 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of metallo-DNA with consecutive thymine-HgII-thymine base pairs explains positive entropy for the metallo base pair formation.
Nucleic Acids Res., 42, 2014
|
|
2RVP
 
 | | Solution structure of DNA Containing Metallo-Base-Pair | | Descriptor: | DNA (5'-D(*TP*AP*AP*TP*AP*TP*AP*CP*TP*TP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*AP*AP*CP*TP*AP*TP*AP*TP*TP*A)-3'), SILVER ION | | Authors: | Dairaku, T, Furuita, K, Sato, H, Sebera, J, Nakashima, K, Kondo, J, Yamanaka, D, Kondo, Y, Okamoto, I, Ono, A, Sychrovsky, V, Kojima, C, Tanaka, Y. | | Deposit date: | 2016-03-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of an Ag(I) -Mediated Cytosine-Cytosine Base Pair within DNA Duplex in Solution with (1) H/(15) N/(109) Ag NMR Spectroscopy.
Chemistry, 22, 2016
|
|
5DZL
 
 | | Crystal structure of the protein human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1 | | Authors: | Huang, Y.H, Russell, A, Gandhi, A.K, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4006 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
3SI8
 
 | | Human DNA polymerase eta - DNA ternary complex with the 5'T of a CPD in the active site (TT2) | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, ... | | Authors: | Biertumpfel, C, Zhao, Y, Kondo, Y, Ramon-Maiques, S, Gregory, M, Lee, J.Y, Masutani, C, Lehmann, A.R, Hanaoka, F, Yang, W. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
4TWT
 
 | | Human TNFa dimer in complex with the semi-synthetic bicyclic peptide M21 | | Descriptor: | (2,4,6-trimethylbenzene-1,3,5-triyl)trimethanol, ALA-CYS-PRO-PRO-CYS-LEU-TRP-GLN-VAL-LEU-CYS-GLY, GLYCEROL, ... | | Authors: | Luzi, S, Kondo, Y, Bernard, E, Stadler, L, Winter, G, Holliger, P. | | Deposit date: | 2014-07-01 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Subunit disassembly and inhibition of TNF alpha by a semi-synthetic bicyclic peptide.
Protein Eng.Des.Sel., 28, 2015
|
|
4QXW
 
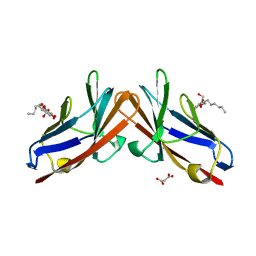 | | Crystal structure of the human CEACAM1 membrane distal amino terminal (N)-domain | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | Authors: | Huang, Y.H, Gandhi, A.K, Russell, A, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2014-07-22 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
4P86
 
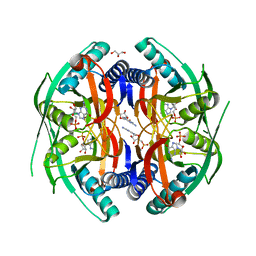 | | Structure of PyrR protein from Bacillus subtilis with GMP | | Descriptor: | Bifunctional protein PyrR, GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4PKD
 
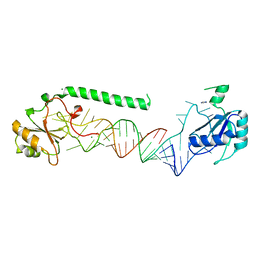 | | U1-70k in complex with U1 snRNA stem-loops 1 and U1-A RRM in complex with stem-loop 2 | | Descriptor: | IMIDAZOLE, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A,U1 small nuclear ribonucleoprotein 70 kDa, ... | | Authors: | Oubridge, C, Kondo, Y, van Roon, A.M, Nagai, K. | | Deposit date: | 2014-05-14 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5' splice site recognition.
Elife, 4, 2015
|
|
4P80
 
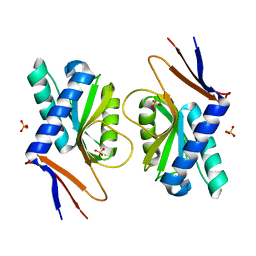 | | Structure of ancestral PyrR protein (AncGREENPyrR) | | Descriptor: | Ancestral PyrR protein (Green), SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-29 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P83
 
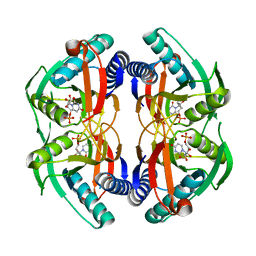 | | Structure of engineered PyrR protein (PURPLE PyrR) | | Descriptor: | Engineered PyrR protein (Purple), URIDINE-5'-MONOPHOSPHATE | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P3K
 
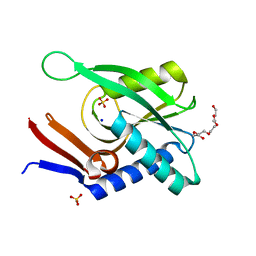 | | Structure of ancestral PyrR protein (PLUMPyrR) | | Descriptor: | Ancestral PyrR protein (Plum), PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-08 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P81
 
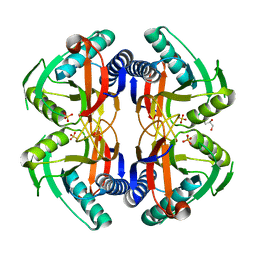 | | Structure of ancestral PyrR protein (AncORANGEPyrR) | | Descriptor: | Ancestral PyrR protein (Orange), GLYCEROL, SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P84
 
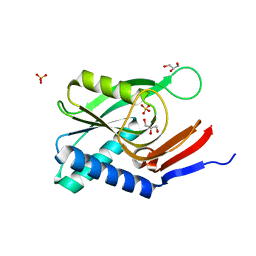 | | Structure of engineered PyrR protein (VIOLET PyrR) | | Descriptor: | Bifunctional protein PyrR, GLYCEROL, SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P82
 
 | | Structure of PyrR protein from Bacillus subtilis | | Descriptor: | Bifunctional protein PyrR, SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
6K53
 
 | | A variant of metagenome-derived GH6 cellobiohydrolase, HmCel6A (P88S/L230F/F414S) | | Descriptor: | CITRATE ANION, GH6 cellobiohydrolase, HMCEL6A, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A hyperthermophilic GH6 cellobiohydrolase (HmCel6A) from a hot spring metagenomic data
To Be Published
|
|
6K52
 
 | | Hyperthermophilic GH6 cellobiohydrolase (HmCel6A) from the microbial flora of a Japanese hot spring | | Descriptor: | ACETATE ION, CALCIUM ION, GH6 cellobiohydrolase, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68000138 Å) | | Cite: | A hyperthermophilic cellobiohydrolase mined from a hot spring metagenomic data
To Be Published
|
|
6K55
 
 | | Inactivated mutant (D140A) of Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with hexasaccharide | | Descriptor: | Glucanase, MAGNESIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | A hyperthermophilic cellobiohydrolase mined from a hot spring metagenomic data
To Be Published
|
|
6K54
 
 | | Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with trisaccharide | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Novel hyperthermophilic cellobiohydrolase II isolated from hot spring microbial community
To Be Published
|
|
