4U5X
 
 | | Structure of plant small GTPase OsRac1 complexed with the non-hydrolyzable GTP analog GMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ohki, I, Kosami, K, Fujiwara, T, Nakagawa, A, Shimamoto, K, Kojima, C. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of the Plant Small GTPase OsRac1 Reveals Its Mode of Binding to NADPH Oxidase
J.Biol.Chem., 289, 2014
|
|
1C4L
 
 | | SOLUTION STRUCTURE OF AN RNA DUPLEX INCLUDING A C-U BASE-PAIR | | Descriptor: | RNA (5'-R(*CP*CP*UP*GP*CP*GP*UP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*UP*CP*AP*GP*G)-3') | | Authors: | Tanaka, Y, Kojima, C, Yamazaki, T, Kodama, T.S, Yasuno, K, Miyashita, S, Ono, A.M, Ono, A.S, Kainosho, M, Kyogoku, Y. | | Deposit date: | 1999-08-30 | | Release date: | 2000-08-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA duplex including a C-U base pair.
Biochemistry, 39, 2000
|
|
7YVW
 
 | | NMR determination of the 2:1 binding motif structure involving cytosine flipping out for the recognition of the CGG/CGG triad DNA | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3') | | Authors: | Furuita, K, Yamada, T, Sakurabayashi, S, Nomura, M, Kojima, C, Nakatani, K. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR determination of the 2:1 binding complex of naphthyridine carbamate dimer (NCD) and CGG/CGG triad in double-stranded DNA.
Nucleic Acids Res., 50, 2022
|
|
1DRN
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DUPLEX CONTAINING DNA/RNA HYBRID REGION, D(GGAGA)R(UGAC)/D(GTCATCTCC) | | Descriptor: | DNA (5'-D(*GP*TP*CP*AP*TP*CP*TP*CP*C)-3'), DNA/RNA (5'-D(*GP*GP*AP*GP*A)-R(P*UP*GP*AP*C)-3') | | Authors: | Nishizaki, T, Iwai, S, Ohkubo, T, Kojima, C, Nakamura, H, Kyogoku, Y, Ohtsuka, E. | | Deposit date: | 1995-11-15 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of DNA duplexes containing a DNA x RNA hybrid region, d(GG)r(AGAU)d(GAC) x d(GTCATCTCC) and d(GGAGA)r(UGAC) x d(GTCATCTCC).
Biochemistry, 35, 1996
|
|
1DHH
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DUPLEX CONTAINING DNA/RNA HYBRID REGION, D(GG)R(AGAU)D(GAC)/D(GTCATCTCC) | | Descriptor: | DNA (5'-D(*GP*TP*CP*AP*TP*CP*TP*CP*C)-3'), DNA/RNA (5'-D(*GP*G)-R(P*AP*GP*AP*U)-D(P*GP*AP*C)-3') | | Authors: | Nishizaki, T, Iwai, S, Ohkubo, T, Kojima, C, Nakamura, H, Kyogoku, Y, Ohtsuka, E. | | Deposit date: | 1995-10-18 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of DNA duplexes containing a DNA x RNA hybrid region, d(GG)r(AGAU)d(GAC) x d(GTCATCTCC) and d(GGAGA)r(UGAC) x d(GTCATCTCC).
Biochemistry, 35, 1996
|
|
8HEW
 
 | | Potato 14-3-3 St14f | | Descriptor: | 14-3-3 protein, StFDL1 peptide | | Authors: | Taoka, K, Kawahara, I, Shinya, S, Harada, K, Muranaka, T, Furuita, K, Nakagawa, A, Fujiwara, T, Tsuji, H, Kojima, C. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Multifunctional chemical inhibitors of the florigen activation complex discovered by structure-based high-throughput screening.
Plant J., 112, 2022
|
|
1UEL
 
 | | Solution structure of ubiquitin-like domain of hHR23B complexed with ubiquitin-interacting motif of proteasome subunit S5a | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, UV excision repair protein RAD23 homolog B | | Authors: | Fujiwara, K, Tenno, T, Jee, J.G, Sugasawa, K, Ohki, I, Kojima, C, Tochio, H, Hiroaki, H, Hanaoka, H, Shirakawa, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-19 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ubiquitin-interacting Motif of S5a Bound to the Ubiquitin-like Domain of HR23B
J.Biol.Chem., 279, 2004
|
|
1UGL
 
 | | Solution structure of S8-SP11 | | Descriptor: | S-locus pollen protein | | Authors: | Mishima, M, Takayama, S, Sasaki, K, Jee, J.G, Kojima, C, Isogai, A, Shirakawa, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-16 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the Male Determinant Factor for Brassica Self-incompatibility
J.Biol.Chem., 278, 2003
|
|
2E30
 
 | |
2ND5
 
 | | Lysine dimethylated FKBP12 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Hattori, Y, Sebera, J, Sychrovsky, V, Furuita, K, Sugiki, T, Ohki, I, Ikegami, T, Kobayashi, N, Tanaka, Y, Fujiwara, T, Kojima, C. | | Deposit date: | 2016-05-05 | | Release date: | 2017-05-17 | | Method: | SOLUTION NMR | | Cite: | NMR Observation of Protein Surface Salt Bridges at Neutral pH
To be Published
|
|
7X8V
 
 | | Cooperative regulation of PBI1 and MAPKs controls WRKY45 transcription factor in rice immunity | | Descriptor: | Os01g0156300 protein | | Authors: | Ichimaru, K, Harada, K, Yamaguchi, K, Shigeta, S, Shimada, K, Ishikawa, K, Inoue, K, Nishio, Y, Yoshimura, S, Inoue, H, Yamashita, E, Fujiwara, T, Nakagawa, A, Kojima, C, Kawasaki, T. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cooperative regulation of PBI1 and MAPKs controls WRKY45 transcription factor in rice immunity.
Nat Commun, 13, 2022
|
|
7XBQ
 
 | |
1X60
 
 | | Solution structure of the peptidoglycan binding domain of B. subtilis cell wall lytic enzyme CwlC | | Descriptor: | Sporulation-specific N-acetylmuramoyl-L-alanine amidase | | Authors: | Mishima, M, Shida, T, Yabuki, K, Kato, K, Sekiguchi, J, Kojima, C. | | Deposit date: | 2005-05-17 | | Release date: | 2005-08-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Peptidoglycan Binding Domain of Bacillus subtilis Cell Wall Lytic Enzyme CwlC: Characterization of the Sporulation-Related Repeats by NMR(,)
Biochemistry, 44, 2005
|
|
1X26
 
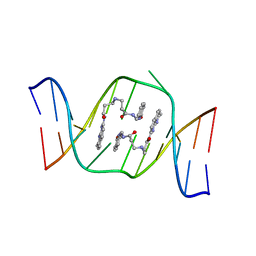 | | Solution structure of the AA-mismatch DNA complexed with naphthyridine-azaquinolone | | Descriptor: | 5'-D(*CP*AP*TP*TP*CP*AP*GP*TP*TP*AP*G)-3', 5'-D(*CP*TP*AP*AP*CP*AP*GP*AP*AP*TP*G)-3', N~3~-{3-[(7-METHYL-1,8-NAPHTHYRIDIN-2-YL)AMINO]-3-OXOPROPYL}-N~1~-[(7-OXO-7,8-DIHYDRO-1,8-NAPHTHYRIDIN-2-YL)METHYL]-BET A-ALANINAMIDE | | Authors: | Nakatani, K, Hagihara, S, Goto, Y, Kobori, A, Hagihara, M, Hayashi, G, Kyo, M, Nomura, M, Mishima, M, Kojima, C. | | Deposit date: | 2005-04-20 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats
Nat.Chem.Biol., 1, 2005
|
|
5GIW
 
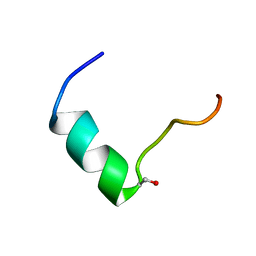 | | Solution NMR structure of Humanin containing a D-isomerized serine residue | | Descriptor: | Humanin | | Authors: | Furuita, K, Sugiki, T, Alsanousi, N, Fujiwara, T, Kojima, C. | | Deposit date: | 2016-06-25 | | Release date: | 2016-07-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and inhibitory effect against amyloid-beta fibrillation of Humanin containing a d-isomerized serine residue
Biochem.Biophys.Res.Commun., 477, 2016
|
|
3AXY
 
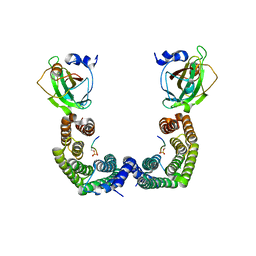 | | Structure of Florigen Activation Complex Consisting of Rice Florigen Hd3a, 14-3-3 Protein GF14 and Rice FD Homolog OsFD1 | | Descriptor: | 14-3-3-like protein GF14-C, Protein HEADING DATE 3A, Rice FD homolog OsFD1 | | Authors: | Ohki, I, Furuita, K, Hayashi, K, Taoka, K, Tsuji, H, Nakagawa, A, Shimamoto, K, Kojima, C. | | Deposit date: | 2011-04-19 | | Release date: | 2011-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen
Nature, 476, 2011
|
|
2RRR
 
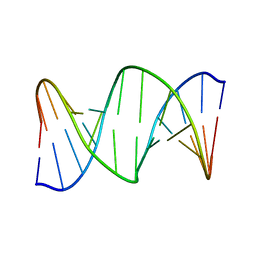 | | DNA oligomer containing ethylene cross-linked cyclic 2'-deoxyuridylate dimer | | Descriptor: | DNA (5'-D(*CP*CP*TP*TP*CP*AP*TP*TP*AP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*TP*AP*AP*TP*GP*AP*AP*GP*G)-3') | | Authors: | Furuita, K, Murata, S, Jee, J.G, Ichikawa, S, Matsuda, A, Kojima, C. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Feature of Bent DNA Recognized by HMGB1
J.Am.Chem.Soc., 133, 2011
|
|
2RUJ
 
 | | Solution structure of MTSL spin-labeled Schizosaccharomyces pombe Sin1 CRIM domain | | Descriptor: | Stress-activated map kinase-interacting protein 1 | | Authors: | Furuita, K, Kataoka, S, Sugiki, T, Kobayashi, N, Ikegami, T, Shiozaki, K, Fujiwara, T, Kojima, C. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Method: | SOLUTION NMR | | Cite: | Utilization of paramagnetic relaxation enhancements for high-resolution NMR structure determination of a soluble loop-rich protein with sparse NOE distance restraints
J.Biomol.Nmr, 61, 2015
|
|
2RR3
 
 | | Solution structure of the complex between human VAP-A MSP domain and human OSBP FFAT motif | | Descriptor: | Oxysterol-binding protein 1, Vesicle-associated membrane protein-associated protein A | | Authors: | Furuita, K, Jee, J, Fukada, H, Mishima, M, Kojima, C. | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Electrostatic interaction between oxysterol-binding protein and VAMP-associated protein A revealed by NMR and mutagenesis studies
J.Biol.Chem., 285, 2010
|
|
2RRQ
 
 | | DNA oligomer containing propylene cross-linked cyclic 2'-deoxyuridylate dimer | | Descriptor: | DNA (5'-D(*CP*CP*TP*TP*CP*AP*(JDT)P*TP*AP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*TP*AP*AP*TP*GP*AP*AP*GP*G)-3') | | Authors: | Furuita, K, Murata, S, Jee, J.G, Ichikawa, S, Matsuda, A, Kojima, C. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Feature of Bent DNA Recognized by HMGB1
J.Am.Chem.Soc., 133, 2011
|
|
2RT8
 
 | | Structure of metallo-dna in solution | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*CP*GP*CP*G)-3'), MERCURY (II) ION | | Authors: | Yamaguchi, H, Sebera, J, Kondo, J, Oda, S, Komuro, T, Kawamura, T, Dairaku, T, Kondo, Y, Okamoto, I, Ono, A, Burda, J.V, Kojima, C, Sychrovsky, V, Tanaka, Y. | | Deposit date: | 2013-06-18 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of metallo-DNA with consecutive thymine-HgII-thymine base pairs explains positive entropy for the metallo base pair formation.
Nucleic Acids Res., 42, 2014
|
|
2RM8
 
 | | The solution structure of phototactic transducer protein HtrII linker region from Natronomonas pharaonis | | Descriptor: | Sensory rhodopsin II transducer | | Authors: | Hayashi, K, Sudo, Y, Jee, J, Mishima, M, Hara, H, Kamo, N, Kojima, C. | | Deposit date: | 2007-10-15 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Phototactic Transducer Protein HtrII Linker Region from Natronomonas pharaonis
Biochemistry, 46, 2007
|
|
2RVK
 
 | |
2RVP
 
 | | Solution structure of DNA Containing Metallo-Base-Pair | | Descriptor: | DNA (5'-D(*TP*AP*AP*TP*AP*TP*AP*CP*TP*TP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*AP*AP*CP*TP*AP*TP*AP*TP*TP*A)-3'), SILVER ION | | Authors: | Dairaku, T, Furuita, K, Sato, H, Sebera, J, Nakashima, K, Kondo, J, Yamanaka, D, Kondo, Y, Okamoto, I, Ono, A, Sychrovsky, V, Kojima, C, Tanaka, Y. | | Deposit date: | 2016-03-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of an Ag(I) -Mediated Cytosine-Cytosine Base Pair within DNA Duplex in Solution with (1) H/(15) N/(109) Ag NMR Spectroscopy.
Chemistry, 22, 2016
|
|
7E3N
 
 | | Crystal structure of Isocitrate dehydrogenase D252N mutant from Trypanosoma brucei in complexed with NADP+, alpha-ketoglutarate, and ca2+ | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], ... | | Authors: | Arai, N, Shiba, T, Inaoka, D.K, Kita, K, Wang, X, Otani, M, Matsushiro, S, Kojima, C. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Isocitrate dehydrogenase from Trypanosoma brucei.
To Be Published
|
|
