5V7P
 
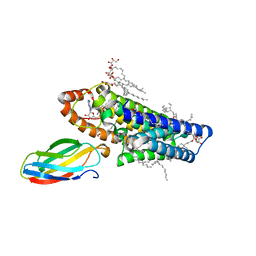 | | Atomic structure of the eukaryotic intramembrane Ras methyltransferase ICMT (isoprenylcysteine carboxyl methyltransferase), in complex with a monobody | | Descriptor: | DECANE, Protein-S-isoprenylcysteine O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Long, S.B, Diver, M.M, Pedi, L, Koide, A, Koide, S. | | Deposit date: | 2017-03-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT.
Nature, 553, 2018
|
|
4IOF
 
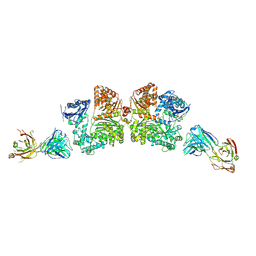 | | Crystal structure analysis of Fab-bound human Insulin Degrading Enzyme (IDE) | | Descriptor: | Fab-bound IDE, heavy chain, light chain, ... | | Authors: | McCord, L.A, Liang, W.G, Hoey, R, Dowdell, E, Koide, A, Koide, S, Tang, W.J. | | Deposit date: | 2013-01-07 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.353 Å) | | Cite: | Conformational states and recognition of amyloidogenic peptides of human insulin-degrading enzyme.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6CPG
 
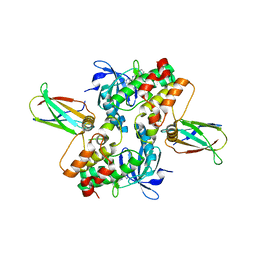 | | Structure of dephosphorylated Aurora A (122-403) in complex with inhibiting monobody and AT9283 in an inactive conformation | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Aurora kinase A, Monobody | | Authors: | Otten, R, Kutter, S, Zorba, A, Padua, R.A.P, Koide, A, Koide, S, Kern, D. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dynamics of human protein kinase Aurora A linked to drug selectivity.
Elife, 7, 2018
|
|
2AF5
 
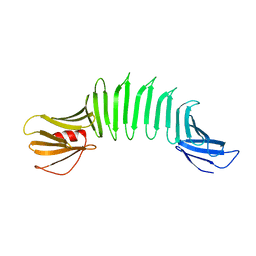 | | 2.5A X-ray Structure of Engineered OspA protein | | Descriptor: | Engineered Outer Surface Protein A (OspA) with the inserted two beta-hairpins | | Authors: | Makabe, K, Mcelheny, D, Tereshko, V, Hilyard, A, Koide, A, Koide, S. | | Deposit date: | 2005-07-25 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structures of peptide self-assembly mimics.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5CWW
 
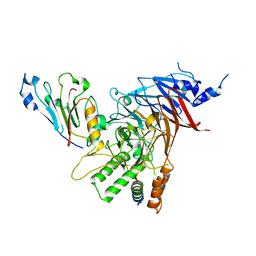 | | Crystal structure of the Chaetomium thermophilum heterotrimeric Nup82 NTD-Nup159 TAIL-Nup145N APD complex | | Descriptor: | Nucleoporin NUP145N, Nucleoporin NUP159, Nucleoporin NUP82 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWV
 
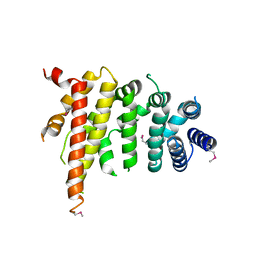 | | Crystal structure of Chaetomium thermophilum Nup192 TAIL domain | | Descriptor: | Nucleoporin NUP192 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.155 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWU
 
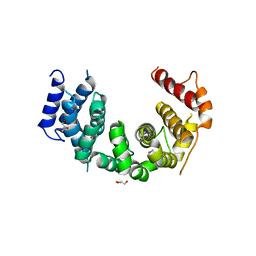 | | Crystal structure of Chaetomium thermophilum Nup188 TAIL domain | | Descriptor: | GLYCEROL, Nucleoporin NUP188 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
8EZG
 
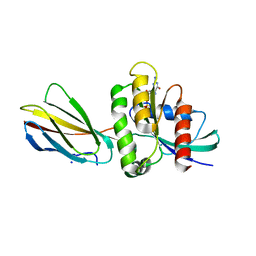 | | Monobody 12D1 bound to KRAS(G12D) | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hattori, T, Glasser, E, Akkapeddi, P, Ketavarapu, G, Teng, K.W, Koide, A, Koide, S. | | Deposit date: | 2022-10-31 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Exploring switch II pocket conformation of KRAS(G12D) with mutant-selective monobody inhibitors.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F0M
 
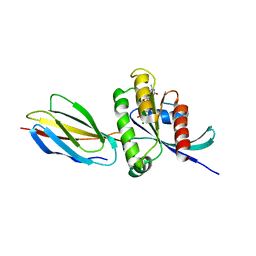 | | Monobody 12D5 bound to KRAS(G12D) | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Hattori, T, Glasser, E, Akkapeddi, P, Ketavarapu, G, Teng, K.W, Koide, A, Koide, S. | | Deposit date: | 2022-11-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Exploring switch II pocket conformation of KRAS(G12D) with mutant-selective monobody inhibitors.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5VG9
 
 | | Structure of the eukaryotic intramembrane Ras methyltransferase ICMT (isoprenylcysteine carboxyl methyltransferase) without a monobody | | Descriptor: | Protein-S-isoprenylcysteine O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Long, S.B, Diver, M.M, Pedi, L, Koide, A, Koide, S. | | Deposit date: | 2017-04-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT.
Nature, 553, 2018
|
|
5DC4
 
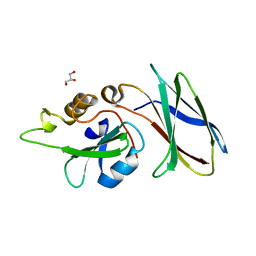 | | CRYSTAL STRUCTURE OF MONOBODY AS25/ABL1 SH2 DOMAIN COMPLEX, CRYSTAL A | | Descriptor: | AS25 monobody, GLYCEROL, Tyrosine-protein kinase ABL1 | | Authors: | Wojcik, J.B, Koide, A, Koide, S. | | Deposit date: | 2015-08-23 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Allosteric Inhibition of Bcr-Abl Kinase by High Affinity Monobody Inhibitors Directed to the Src Homology 2 (SH2)-Kinase Interface.
J.Biol.Chem., 291, 2016
|
|
5DC9
 
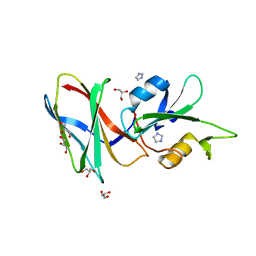 | | CRYSTAL STRUCTURE OF MONOBODY AS25/ABL1 SH2 DOMAIN COMPLEX, CRYSTAL B | | Descriptor: | AS25 monobody, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wojcik, J.B, Koide, A, Koide, S. | | Deposit date: | 2015-08-23 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Allosteric Inhibition of Bcr-Abl Kinase by High Affinity Monobody Inhibitors Directed to the Src Homology 2 (SH2)-Kinase Interface.
J.Biol.Chem., 291, 2016
|
|
5A40
 
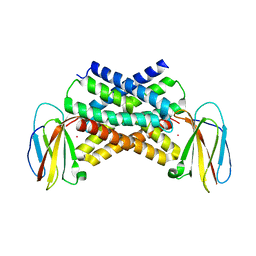 | | Crystal structure of a dual topology fluoride ion channel. | | Descriptor: | MERCURY (II) ION, MONOBODIES, PUTATIVE FLUORIDE ION TRANSPORTER CRCB | | Authors: | Stockbridge, R.B, Kolmakova-Partensky, L, Shane, T, Koide, A, Koide, S, Miller, C, Newstead, S. | | Deposit date: | 2015-06-04 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structures of a Double-Barrelled Fluoride Ion Channel.
Nature, 525, 2015
|
|
5A43
 
 | | Crystal structure of a dual topology fluoride ion channel. | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, FLUORIDE ION, MONOBODIES, ... | | Authors: | Stockbridge, R.B, Kolmakova-Partensky, L, Shane, T, Koide, A, Koide, S, Miller, C, Newstead, S. | | Deposit date: | 2015-06-04 | | Release date: | 2015-09-02 | | Last modified: | 2015-09-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal Structures of a Double-Barrelled Fluoride Ion Channel.
Nature, 525, 2015
|
|
5G15
 
 | | Structure Aurora A (122-403) bound to activating monobody Mb1 and AMPPCP | | Descriptor: | AURORA A KINASE, MAGNESIUM ION, MB1 MONOBODY, ... | | Authors: | Zorba, A, Kutter, S, Kern, D, Koide, S, Koide, A. | | Deposit date: | 2016-03-23 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Allosteric modulation of a human protein kinase with monobodies.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5E95
 
 | | Crystal Structure of Mb(NS1)/H-Ras Complex | | Descriptor: | GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eguchi, R.R, Sha, F, Gupta, A, Koide, A, Koide, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Inhibition of RAS function through targeting an allosteric regulatory site.
Nat. Chem. Biol., 13, 2017
|
|
2KIR
 
 | | Solution structure of a designer toxin, mokatoxin-1 | | Descriptor: | Designer toxin | | Authors: | Biancalana, M, Koide, A, Takacs, Z, Goldstein, S, Koide, S. | | Deposit date: | 2009-05-07 | | Release date: | 2009-12-29 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A designer ligand specific for Kv1.3 channels from a scorpion neurotoxin-based library.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2OCF
 
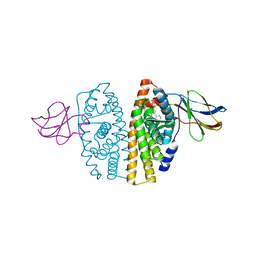 | | Human estrogen receptor alpha ligand-binding domain in complex with estradiol and the E2#23 FN3 monobody | | Descriptor: | ESTRADIOL, Estrogen receptor, Fibronectin | | Authors: | Rajan, S.S, Kuruvilla, S.M, Sharma, S.K, Kim, Y, Huang, J, Koide, A, Koide, S, Joachimiak, A, Greene, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6D0N
 
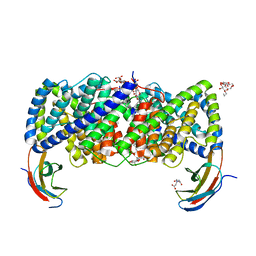 | | Crystal structure of a CLC-type fluoride/proton antiporter, V319G mutant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6D0K
 
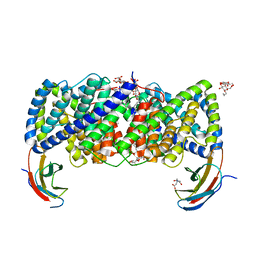 | | Crystal structure of a CLC-type fluoride/proton antiporter, E118Q mutant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6D0J
 
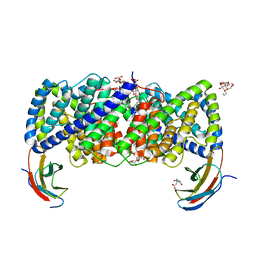 | | Crystal structure of a CLC-type fluoride/proton antiporter | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7LO8
 
 | | NorA in complex with Fab36 | | Descriptor: | Fab36 Heavy Chain, Fab36 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
7LO7
 
 | | NorA in complex with Fab25 | | Descriptor: | Fab25 Heavy Chain, Fab25 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
3UYO
 
 | |
2Z8G
 
 | | Aspergillus niger ATCC9642 isopullulanase complexed with isopanose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Mizuno, M, Koide, A, Yamamura, A, Akeboshi, H, Yoshida, H, Kamitori, S, Sakano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2007-09-05 | | Release date: | 2007-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49
J.Mol.Biol., 376, 2008
|
|
