2BNH
 
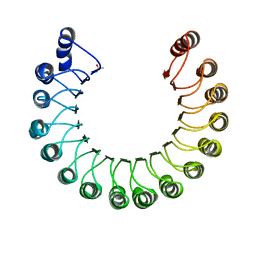 | | PORCINE RIBONUCLEASE INHIBITOR | | Descriptor: | RIBONUCLEASE INHIBITOR | | Authors: | Kobe, B, Deisenhofer, J. | | Deposit date: | 1996-06-29 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A.
J.Mol.Biol., 264, 1996
|
|
1DFJ
 
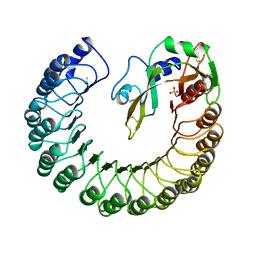 | |
1PHZ
 
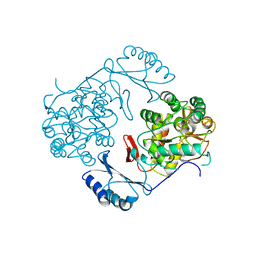 | | STRUCTURE OF PHOSPHORYLATED PHENYLALANINE HYDROXYLASE | | Descriptor: | FE (III) ION, PROTEIN (PHENYLALANINE HYDROXYLASE) | | Authors: | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | Deposit date: | 1998-11-11 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
2PHM
 
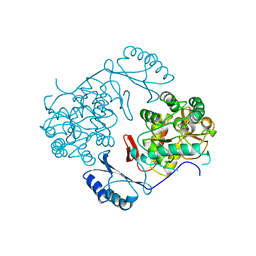 | | STRUCTURE OF PHENYLALANINE HYDROXYLASE DEPHOSPHORYLATED | | Descriptor: | FE (III) ION, PROTEIN (PHENYLALANINE-4-HYDROXYLASE) | | Authors: | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | Deposit date: | 1998-11-11 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
1KOB
 
 | | TWITCHIN KINASE FRAGMENT (APLYSIA), AUTOREGULATED PROTEIN KINASE DOMAIN | | Descriptor: | TWITCHIN, VALINE | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
1KOA
 
 | | TWITCHIN KINASE FRAGMENT (C.ELEGANS), AUTOREGULATED PROTEIN KINASE AND IMMUNOGLOBULIN DOMAINS | | Descriptor: | TWITCHIN | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
1MG1
 
 | | HTLV-1 GP21 ECTODOMAIN/MALTOSE-BINDING PROTEIN CHIMERA | | Descriptor: | CHLORIDE ION, PROTEIN (HTLV-1 GP21 ECTODOMAIN/MALTOSE-BINDING PROTEIN CHIMERA), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kobe, B, Center, R.J, Kemp, B.E, Poumbourios, P. | | Deposit date: | 1999-03-01 | | Release date: | 1999-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1IAL
 
 | | IMPORTIN ALPHA, MOUSE | | Descriptor: | IMPORTIN ALPHA | | Authors: | Kobe, B. | | Deposit date: | 1999-01-12 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha.
Nat.Struct.Biol., 6, 1999
|
|
1EJL
 
 | |
4ZM1
 
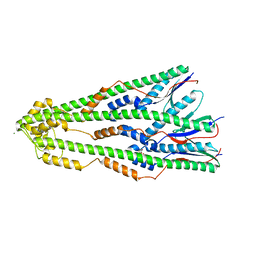 | | Shigella flexneri lipopolysaccharide O-antigen chain-length regulator WzzBSF - wild type | | Descriptor: | CITRIC ACID, Chain length determinant protein, MAGNESIUM ION | | Authors: | Ericsson, D.J, Chang, C.-W, Lonhienne, T, Casey, L, Benning, F, Kobe, B, Tran, E.N.H, Morona, R. | | Deposit date: | 2015-05-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Biochemical Analysis of a Single Amino-Acid Mutant of WzzBSF That Alters Lipopolysaccharide O-Antigen Chain Length in Shigella flexneri.
Plos One, 10, 2015
|
|
4UTP
 
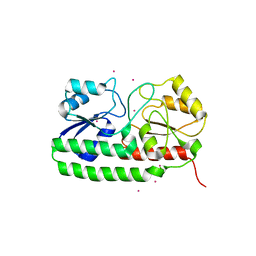 | | Crystal structure of pneumococcal surface antigen PsaA in the Cd- bound, closed state | | Descriptor: | CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
7SHJ
 
 | |
8STB
 
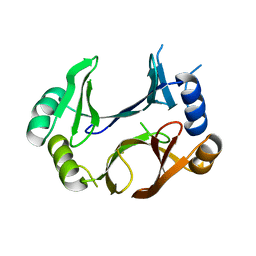 | | The structure of abxF, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX | | Descriptor: | CHLORIDE ION, GLYCEROL, Glyoxalase, ... | | Authors: | Luo, Z, Jia, X, Yan, X, Qu, X, Kobe, B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The crystal structure of abxF, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX.
To Be Published
|
|
4UTO
 
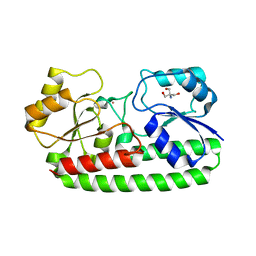 | | Crystal structure of pneumococcal surface antigen PsaA D280N in the Cd-bound, open state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
7RX1
 
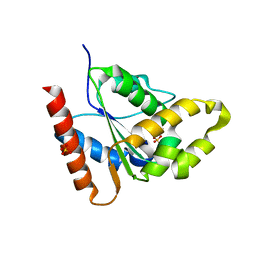 | |
7S2Z
 
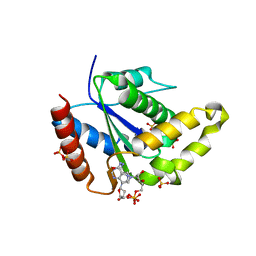 | |
8JZM
 
 | | The inhibitor of Toll-like receptor signaling o-vanillin binds covalently to MAL/TIRAP Lys-210 | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Rahaman, M.H, Jia, X, Maxwell, M.J, Mobli, M, Kobe, B. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | o-Vanillin binds covalently to MAL/TIRAP Lys-210 but independently inhibits TLR2.
J Enzyme Inhib Med Chem, 39, 2024
|
|
5WEG
 
 | | Crystal Structure of UDP-glucose pyrophosphorylase from Sugarcane | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UTP--glucose-1-phosphate uridylyltransferase | | Authors: | Cotrim, C.A, Soares, J.S.M, Kobe, B, Menossi, M. | | Deposit date: | 2017-07-10 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and insights into the oligomeric state of UDP-glucose pyrophosphorylase from sugarcane.
PLoS ONE, 13, 2018
|
|
2V1O
 
 | | Crystal structure of N-terminal domain of acyl-CoA thioesterase 7 | | Descriptor: | COENZYME A, CYTOSOLIC ACYL COENZYME A THIOESTER HYDROLASE | | Authors: | Forwood, J.K, Thakur, A.S, Guncar, G, Marfori, M, Mouradov, D, Meng, W.N, Robinson, J, Huber, T, Kellie, S, Martin, J.L, Hume, D.A, Kobe, B. | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Recruitment of Tandem Hotdog Domains in Acyl-Coa Thioesterase 7 and its Role in Inflammation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3EA5
 
 | | Kap95p Binding Induces the Switch Loops of RanGDP to adopt the GTP-bound Conformation: Implications for Nuclear Import Complex Assembly Dynamics | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, Importin subunit beta-1, ... | | Authors: | Forwood, J.K, Lonhienne, J.K, Guncar, G, Stewart, M, Marfori, M, Kobe, B. | | Deposit date: | 2008-08-24 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kap95p binding induces the switch loops of RanGDP to adopt the GTP-bound conformation: implications for nuclear import complex assembly dynamics.
J.Mol.Biol., 383, 2008
|
|
4ZM5
 
 | | Shigella flexneri lipopolysaccharide O-antigen chain-length regulator WzzBSF - A107P mutant | | Descriptor: | CHLORIDE ION, Chain length determinant protein, MAGNESIUM ION | | Authors: | Ericsson, D.J, Chang, C.-W, Lonhienne, T, Casey, L, Benning, F, Kobe, B, Tran, E.N.H, Morona, R. | | Deposit date: | 2015-05-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural and Biochemical Analysis of a Single Amino-Acid Mutant of WzzBSF That Alters Lipopolysaccharide O-Antigen Chain Length in Shigella flexneri.
Plos One, 10, 2015
|
|
7BER
 
 | | SFX structure of the MyD88 TIR domain higher-order assembly (solved, rebuilt and refined using an identical protocol to the MicroED structure of the MyD88 TIR domain higher-order assembly) | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
7BEQ
 
 | | MicroED structure of the MyD88 TIR domain higher-order assembly | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
7RTC
 
 | |
6C80
 
 | | Crystal structure of a flax cytokinin oxidase | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wan, L, Williams, S, Kobe, B. | | Deposit date: | 2018-01-23 | | Release date: | 2018-06-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and functional insights into the modulation of the activity of a flax cytokinin oxidase by flax rust effector AvrL567-A.
Mol. Plant Pathol., 20, 2019
|
|
