6GDC
 
 | | Human Carbonic Anhydrase II in complex with Benzenesulfonamide | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, MERCURY (II) ION, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.079 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
1XL5
 
 | | HIV-1 Protease in complex with amidhyroxysulfone | | Descriptor: | CHLORIDE ION, N-{(1S)-1-(3-BROMOBENZYL)-4-[(4-BROMOPHENYL)SULFONYL]-6-METHYL-2-OXOHEPTYL}-2-(2,6-DIMETHYLPHENOXY)ACETAMIDE, PROTEASE RETROPEPSIN | | Authors: | Boettcher, J, Specker, E, Heine, A, Klebe, G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | An Old Target Revisited: Two New Privileged Skeletons and an Unexpected Binding Mode For HIV-Protease Inhibitors
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
6GM9
 
 | | Human Carbonic Anhydrase II in complex with 4-Methylbenzenesulfonamide | | Descriptor: | 4-methylbenzenesulfonamide, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-05-24 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.089 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
7AZV
 
 | |
7AXY
 
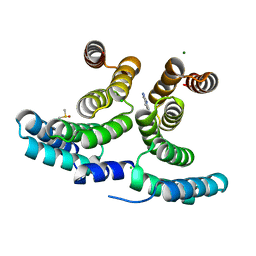 | |
7AYW
 
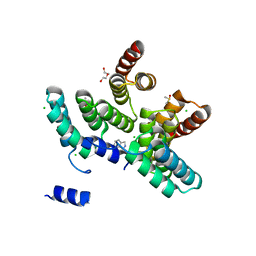 | |
7B1U
 
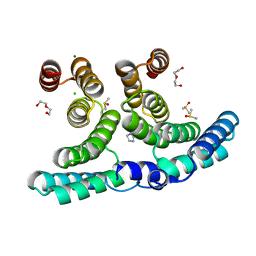 | | Crystal structure of a shortened IpgC variant in complex with 3-(3,4-difluorophenyl)-1H,4H,6H,7H-imidazo[2,1-c][1,2,4]triazine | | Descriptor: | 3-[3,4-bis(fluoranyl)phenyl]-1,4,6,7-tetrahydroimidazo[2,1-c][1,2,4]triazine, CHLORIDE ION, Chaperone protein IpgC, ... | | Authors: | Gardonyi, M, Heine, A, Klebe, G. | | Deposit date: | 2020-11-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of a shortened IpgC variant in complex with 3-(3,4-difluorophenyl)-1H,4H,6H,7H-imidazo[2,1-c][1,2,4]triazine
To be published
|
|
7AC9
 
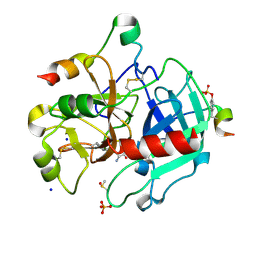 | | Thrombin in complex with D-arginine (j77) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-ARGININE, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-09-10 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.393 Å) | | Cite: | Thrombin in complex with D-arginine (j77)
To be published
|
|
5LPQ
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex (space group P21) with 6-amino-4-(2-((3aR,4R,6R,6aR)-6-methoxy-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)ethyl)-2-(methylamino)-1H-imidazo[4,5-g]quinazolin-8(7H)-one | | Descriptor: | 4-[2-[(3~{a}~{R},4~{R},6~{R},6~{a}~{R})-4-methoxy-2,2-dimethyl-3~{a},4,6,6~{a}-tetrahydrofuro[3,4-d][1,3]dioxol-6-yl]ethyl]-6-azanyl-2-(methylamino)-1,7-dihydroimidazo[4,5-g]quinazolin-8-one, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2016-08-14 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Carbohydrate-based Inhibitors targeting the Ribose-34 pocket of Z.mobilis TGT and changing the oligomeric state of the homodimer
To be published
|
|
6G4L
 
 | | 17beta-hydroxysteroid Dehydrogenase Type 14 Mutant Y253A in Complex With a Non-steroidal Inhibitor | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, DIMETHYL SULFOXIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Badran, M, Klebe, G, Heine, A, Marchais-Oberwinkler, S. | | Deposit date: | 2018-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Mutational and structural studies uncover crucial amino acids determining activity and stability of 17beta-HSD14
J.Steroid Biochem.Mol.Biol., 2019
|
|
5LPT
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex (space group P21) with 6-amino-2-(methylamino)-4-(2-((2R,3R,4S,5R,6S)-3,4,5,6-tetramethoxytetrahydro-2H-pyran-2-yl)ethyl)-1H-imidazo[4,5-g]quinazolin-8(7H)-one | | Descriptor: | 6-azanyl-2-(methylamino)-4-[2-[(2~{R},3~{R},4~{S},5~{R},6~{S})-3,4,5,6-tetramethoxyoxan-2-yl]ethyl]-1,7-dihydroimidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2016-08-14 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Carbohydrate-based Inhibitors targeting the Ribose-34 pocket of Z.mobilis TGT and changing the oligomeric state of the homodimer
To be published
|
|
6H7C
 
 | |
6GTB
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with FB211 | | Descriptor: | 1,2-ETHANEDIOL, 17-beta-hydroxysteroid dehydrogenase 14, 3-[6-(3-hydroxyphenyl)pyridin-2-yl]benzoic acid, ... | | Authors: | Bertoletti, N, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2018-06-18 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | X-ray Crystallographic Fragment screening and Hit Optimization
To Be Published
|
|
1XL2
 
 | | HIV-1 Protease in complex with pyrrolidinmethanamine | | Descriptor: | CHLORIDE ION, GLYCEROL, N-BENZYL-2-(2,6-DIMETHYLPHENOXY)-N-[((3R,4S)-4-{[ISOBUTYL(PHENYLSULFONYL)AMINO]METHYL}PYRROLIDIN-3-YL)METHYL]ACETAMIDE, ... | | Authors: | Boettcher, J, Specker, E, Heine, A, Klebe, G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Old Target Revisited: Two New Privileged Skeletons and an Unexpected Binding Mode For HIV-Protease Inhibitors
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
7APM
 
 | | tRNA-guanine transglycosylase H319C mutant spin-labeled with MTSL. | | Descriptor: | CHLORIDE ION, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2020-10-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unraveling a Ligand-Induced Twist of a Homodimeric Enzyme by Pulsed Electron-Electron Double Resonance.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5LPO
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex with 6-amino-2-(methylamino)-4-(2-((2R,3R,4R,5R)-3,4,5-trimethoxytetrahydrofuran-2-yl)ethyl)-1H-imidazo[4,5-g]quinazolin-8(7H)-one | | Descriptor: | 1,2-ETHANEDIOL, 6-amino-2-(methylamino)-4-(2-((2R,3R,4R,5R)-3,4,5-trimethoxytetrahydrofuran-2-yl)ethyl)-1H-imidazo[4,5-g]quinazolin-8(7H)-one, GLYCEROL, ... | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2016-08-14 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Carbohydrate-based Inhibitors targeting the Ribose-34 pocket of Z.mobilis TGT and changing the oligomeric state of the homodimer
To be published
|
|
4PUN
 
 | | tRNA-Guanine Transglycosylase (TGT) Apo-Structure pH 7.8 | | Descriptor: | DIMETHYL SULFOXIDE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Chasing Protons: How Isothermal Titration Calorimetry, Mutagenesis, and pKa Calculations Trace the Locus of Charge in Ligand Binding to a tRNA-Binding Enzyme.
J.Med.Chem., 57, 2014
|
|
5L8P
 
 | | Thermolysin in complex with JC114 (PEG400 cryo protectant) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, N~2~-[(R)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-N-[(2S)-2,3,3-trimethylbutyl]-L-leucinamide, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-06-08 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Rational Design of Thermodynamic and Kinetic Binding Profiles by Optimizing Surface Water Networks Coating Protein-Bound Ligands.
J. Med. Chem., 59, 2016
|
|
6RM1
 
 | | Human Carbonic Anhydrase II in complex with fragment. | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 3-methylbenzohydrazide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Proof-of-Concept Fragment Screening of a Hit-Validated 96-Compounds Library against Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
6RMP
 
 | | Human Carbonic Anhydrase II in complex with fragment. | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 3-(dimethylamino)benzohydrazide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | A Proof-of-Concept Fragment Screening of a Hit-Validated 96-Compounds Library against Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
5L3U
 
 | | Thermolysin in complex with JC149 (MPD cryo protectant) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, N~2~-[(R)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-N-[(2R)-2,3,3-trimethylbutyl]-L-leucinamide, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Rational Design of Thermodynamic and Kinetic Binding Profiles by Optimizing Surface Water Networks Coating Protein-Bound Ligands.
J. Med. Chem., 59, 2016
|
|
5LCP
 
 | |
5LCT
 
 | |
5LCU
 
 | |
6SDP
 
 | |
