2XB5
 
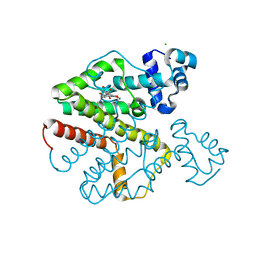 | | Tet repressor (class D) in complex with 7-Iodotetracycline | | Descriptor: | 7-IODOTETRACYCLINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kisker, C, Saenger, W, Hinrichs, W. | | Deposit date: | 2010-04-05 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance.
Science, 264, 1994
|
|
1SOX
 
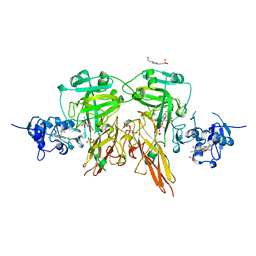 | | SULFITE OXIDASE FROM CHICKEN LIVER | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MOLYBDENUM ATOM, ... | | Authors: | Kisker, C, Schindelin, H, Rees, D.C. | | Deposit date: | 1997-12-31 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of sulfite oxidase deficiency from the structure of sulfite oxidase.
Cell(Cambridge,Mass.), 91, 1997
|
|
1THJ
 
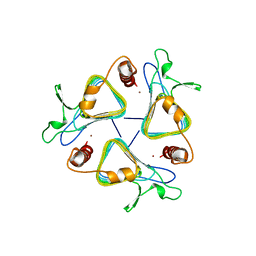 | | CARBONIC ANHYDRASE FROM METHANOSARCINA | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Kisker, C, Schindelin, H, Rees, D.C. | | Deposit date: | 1996-04-02 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila.
EMBO J., 15, 1996
|
|
5G2O
 
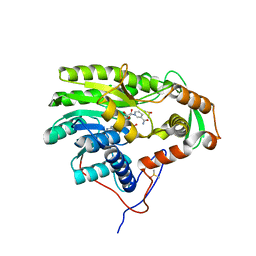 | | Yersinia pestis FabV variant T276A | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-11 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia Pestis Fabv Enoyl-Acp Reductase.
Biochemistry, 55, 2016
|
|
4V98
 
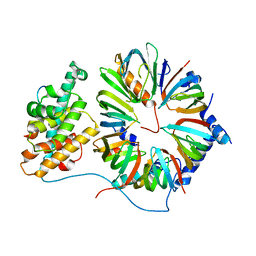 | | The 8S snRNP Assembly Intermediate | | Descriptor: | CG10419, Icln, LD23602p, ... | | Authors: | Grimm, C, Pelz, J.P, Schindelin, H, Diederichs, K, Kuper, J, Kisker, C. | | Deposit date: | 2012-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Assembly Chaperone- Mediated snRNP Formation.
Mol.Cell, 49, 2013
|
|
8REV
 
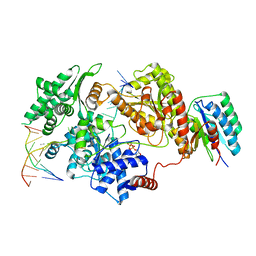 | | Structure of XPD stalled at a Y-fork DNA containing a interstrand crosslink | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase CHL1, DNA (46-MER), ... | | Authors: | Kuper, J, Hove, T, Kisker, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | XPD stalled on cross-linked DNA provides insight into damage verification.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7AD8
 
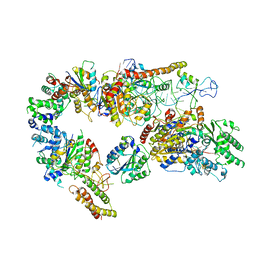 | | Core TFIIH-XPA-DNA complex with modelled p62 subunit | | Descriptor: | DNA (49-MER), DNA repair protein complementing XP-A cells, General transcription and DNA repair factor IIH helicase subunit XPB, ... | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2020-09-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The TFIIH subunits p44/p62 act as a damage sensor during nucleotide excision repair.
Nucleic Acids Res., 48, 2020
|
|
8P1P
 
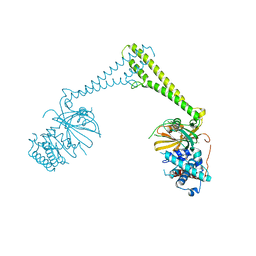 | | USP28 in complex with AZ1 | | Descriptor: | 2-[[5-bromanyl-2-[[4-fluoranyl-3-(trifluoromethyl)phenyl]methoxy]phenyl]methylamino]ethanol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sauer, F, Karal-Nair, R, Kisker, C. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis for the bi-specificity of USP25 and USP28 inhibitors.
Embo Rep., 25, 2024
|
|
1QRE
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | BICARBONATE ION, CARBONIC ANHYDRASE, COBALT (II) ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRL
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | BICARBONATE ION, CARBONIC ANHYDRASE, ZINC ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1Q51
 
 | | Crystal Structure of Mycobacterium tuberculosis MenB in Complex with Acetoacetyl-Coenzyme A, a Key Enzyme in Vitamin K2 Biosynthesis | | Descriptor: | ACETOACETYL-COENZYME A, menB | | Authors: | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-05 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
1Q52
 
 | | Crystal Structure of Mycobacterium tuberculosis MenB, a Key Enzyme in Vitamin K2 Biosynthesis | | Descriptor: | menB | | Authors: | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-05 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
8P1Q
 
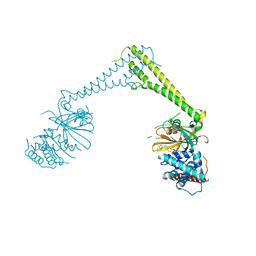 | | USP28 in complex with FT206 | | Descriptor: | 3-azanyl-N-[(2S)-6-[(1S,5R)-3,8-diazabicyclo[3.2.1]octan-3-yl]-1,2,3,4-tetrahydronaphthalen-2-yl]-6-methyl-thieno[2,3-b]pyridine-2-carboxamide, DIMETHYL SULFOXIDE, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Sauer, F, Karal Nair, R, Kisker, C. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for the bi-specificity of USP25 and USP28 inhibitors.
Embo Rep., 25, 2024
|
|
8P19
 
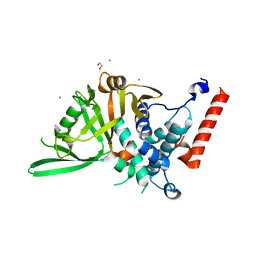 | | USP28 USP domain apo | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Sauer, F, Karal Nair, R, Kisker, C. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the bi-specificity of USP25 and USP28 inhibitors.
Embo Rep., 25, 2024
|
|
8P14
 
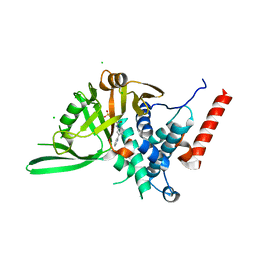 | | USP28 USP domain in complex with Vismodegib | | Descriptor: | 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Sauer, F, Karal Nair, R, Kisker, C. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for the bi-specificity of USP25 and USP28 inhibitors.
Embo Rep., 25, 2024
|
|
3T88
 
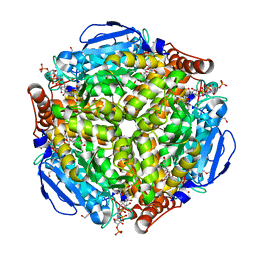 | | Crystal structure of Escherichia coli MenB in complex with substrate analogue, OSB-NCoA | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
3T8A
 
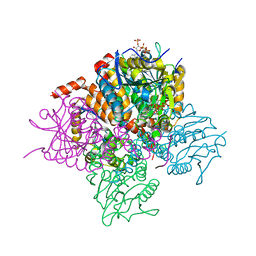 | | Crystal structure of Mycobacterium tuberculosis MenB in complex with substrate analogue, OSB-NCoA | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, o-succinylbenzoyl-N-coenzyme A | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
2X23
 
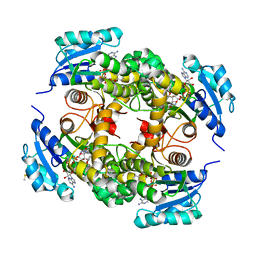 | | crystal structure of M. tuberculosis InhA inhibited by PT70 | | Descriptor: | 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, DIMETHYL SULFOXIDE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], ... | | Authors: | Luckner, S.R, Liu, N, am Ende, C.W, Tonge, P.J, Kisker, C. | | Deposit date: | 2010-01-10 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | A Slow, Tight Binding Inhibitor of Inha, the Enoyl-Acyl Carrier Protein Reductase from Mycobacterium Tuberculosis.
J.Biol.Chem., 285, 2010
|
|
2X22
 
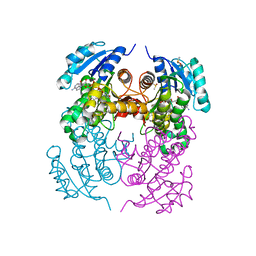 | | crystal structure of M. tuberculosis InhA inhibited by PT70 | | Descriptor: | 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, DIMETHYL SULFOXIDE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], ... | | Authors: | Luckner, S.R, Liu, N, am Ende, C.W, Tonge, P.J, Kisker, C. | | Deposit date: | 2010-01-10 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Slow, Tight Binding Inhibitor of Inha, the Enoyl-Acyl Carrier Protein Reductase from Mycobacterium Tuberculosis.
J.Biol.Chem., 285, 2010
|
|
3T89
 
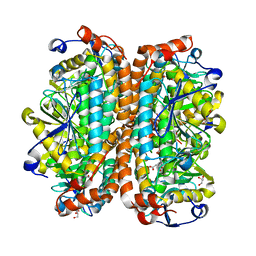 | | Crystal structure of Escherichia coli MenB, the 1,4-dihydroxy-2-naphthoyl-CoA synthase in vitamin K2 biosynthesis | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, GLYCEROL, MALONATE ION | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
4PN7
 
 | |
3T8B
 
 | | Crystal structure of Mycobacterium tuberculosis MenB with altered hexameric assembly | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, GLYCEROL | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
5LY9
 
 | | Structure of MITat 1.1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Variant surface glycoprotein MITAT 1.1, ... | | Authors: | Schaefer, C, Bartossek, T, Jones, N, Kuper, J, Kisker, C, Engstler, M. | | Deposit date: | 2016-09-26 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the shielding function of the dynamic trypanosome variant surface glycoprotein coat.
Nat Microbiol, 2, 2017
|
|
6FDU
 
 | | Structure of Chlamydia trachomatis effector protein Cdu1 bound to Compound 3 | | Descriptor: | (2~{S},3~{S})-2-[[(2~{S})-2-[3,5-bis(chloranyl)phenyl]-2-(dimethylamino)ethanoyl]amino]-~{N}-[[2-(iminomethyl)pyrimidin-4-yl]methyl]-3-methyl-pentanamide, CHLORIDE ION, Deubiquitinase and deneddylase Dub1 | | Authors: | Ramirez, Y, Kisker, C, Altmann, E. | | Deposit date: | 2017-12-26 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Substrate Recognition and Covalent Inhibition of Cdu1 from Chlamydia trachomatis.
ChemMedChem, 13, 2018
|
|
6FDQ
 
 | |
