5FAP
 
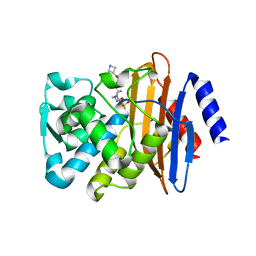 | | CTX-M-15 in complex with FPI-1602 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-6-[(azetidin-3-ylcarbonylamino)carbamoyl]-1-methanoyl-piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FA7
 
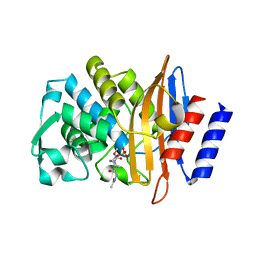 | | CTX-M-15 in complex with FPI-1523 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-6-(acetamidocarbamoyl)-1-methanoyl-piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FAO
 
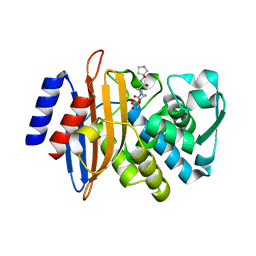 | | CTX-M-15 in complex with FPI-1465 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-1-methanoyl-6-[[(3~{S})-pyrrolidin-3-yl]oxycarbamoyl]piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FAT
 
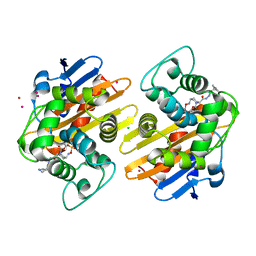 | | OXA-48 in complex with FPI-1602 | | Descriptor: | Beta-lactamase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-12 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FAQ
 
 | | OXA-48 in complex with FPI-1465 | | Descriptor: | Beta-lactamase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FAS
 
 | | OXA-48 in complex with FPI-1523 | | Descriptor: | Beta-lactamase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5VQZ
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N, Y181C) Variant in Complex with 2-chloro-N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacetamide (JLJ686), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Buckingham, A.B, Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2WZR
 
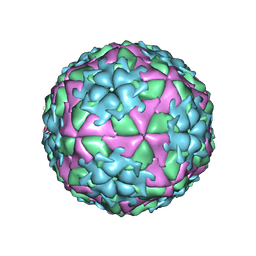 | | The Structure of Foot and Mouth Disease Virus Serotype SAT1 | | Descriptor: | POLYPROTEIN | | Authors: | Adams, P, Lea, S, Newman, J, Blakemore, W, King, A, Stuart, D, Fry, E. | | Deposit date: | 2009-12-02 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Foot-and-Mouth Disease Virus Serotype Sat1.
To be Published
|
|
5KVL
 
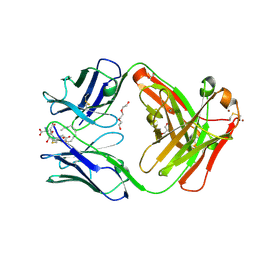 | | Humanized 10G4 anti-leukotriene C4 antibody Fab fragment in complex with leukotriene C4 | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-( 2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Huxford, T, King, A, Fleming, J.K, Wojciak, J.M. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Humanized 10G4 anti-leukotriene C4 antibody Fab fragment in complex with leukotriene C4
To Be Published
|
|
1QQP
 
 | | FOOT-AND-MOUTH DISEASE VIRUS/ OLIGOSACCHARIDE RECEPTOR COMPLEX. | | Descriptor: | 2-O-sulfo-alpha-L-gulopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-gulopyranuronic acid, PROTEIN (GENOME POLYPROTEIN) | | Authors: | Fry, E.E, Lea, S.M, Jackson, T, Newman, J.W.I, Ellard, F.M, Blakemore, W.E, Abu-Ghazaleh, R, Samuel, A, King, A.M.Q, Stuart, D.I. | | Deposit date: | 1999-05-20 | | Release date: | 1999-06-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex.
EMBO J., 18, 1999
|
|
3IPS
 
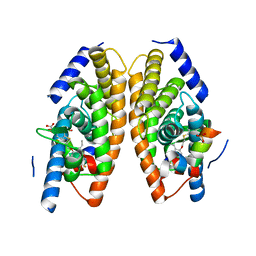 | | X-ray structure of benzisoxazole synthetic agonist bound to the LXR-alpha | | Descriptor: | Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, SULFATE ION, ... | | Authors: | Fradera, X, Vu, D, Nimz, O, Skene, R, Hosfield, D, Wijnands, R, Cooke, A.J, Haunso, A, King, A, Bennet, D.J, McGuire, R, Uitdehaag, J.C.M. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | X-ray structures of the LXRalpha LBD in its homodimeric form and implications for heterodimer signaling.
J.Mol.Biol., 399, 2010
|
|
3IPU
 
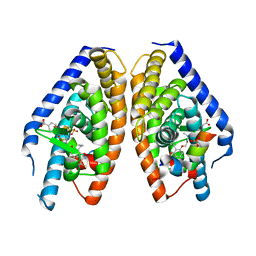 | | X-ray structure of benzisoxazole urea synthetic agonist bound to the LXR-alpha | | Descriptor: | 4-{[methyl(3-{[7-propyl-3-(trifluoromethyl)-1,2-benzisoxazol-6-yl]oxy}propyl)carbamoyl]amino}benzoic acid, Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, ... | | Authors: | Fradera, X, Vu, D, Nimz, O, Skene, R, Hosfield, D, Wijnands, R, Cooke, A.J, Haunso, A, King, A, Bennet, D.J, McGuire, R, Uitdehaag, J.C.M. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of the LXRalpha LBD in its homodimeric form and implications for heterodimer signaling.
J.Mol.Biol., 399, 2010
|
|
3IPQ
 
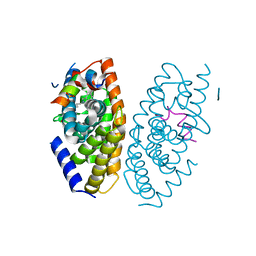 | | X-ray structure of GW3965 synthetic agonist bound to the LXR-alpha | | Descriptor: | Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, SULFATE ION, ... | | Authors: | Fradera, X, Vu, D, Nimz, O, Skene, R, Hosfield, D, Wijnands, R, Cooke, A.J, Haunso, A, King, A, Bennet, D.J, McGuire, R, Uitdehaag, J.C.M. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the LXRalpha LBD in its homodimeric form and implications for heterodimer signaling.
J.Mol.Biol., 399, 2010
|
|
1OOP
 
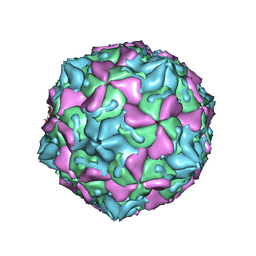 | | The Crystal Structure of Swine Vesicular Disease Virus | | Descriptor: | Coat protein VP1, Coat protein VP2, Coat protein VP3, ... | | Authors: | Fry, E.E, Knowles, N.J, Newman, J.W.I, Wilsden, G, Rao, Z, King, A.M.Q, Stuart, D.I. | | Deposit date: | 2003-03-04 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Swine Vesicular Disease Virus and Implications for Host Adaptation
J.Virol., 77, 2003
|
|
1ZBE
 
 | | Foot-and Mouth Disease Virus Serotype A1061 | | Descriptor: | Coat protein VP1, Coat protein VP2, Coat protein VP3, ... | | Authors: | Fry, E.E, Newman, J.W, Curry, S, Najjam, S, Jackson, T, Blakemore, W, Lea, S.M, Miller, L, Burman, A, King, A.M, Stuart, D.I. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Foot-and-mouth disease virus serotype A1061 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation.
J.Gen.Virol., 86, 2005
|
|
1ZBA
 
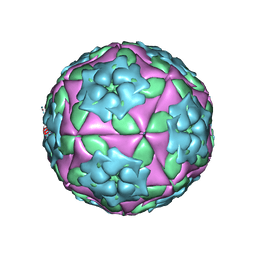 | | Foot-and-Mouth Disease virus serotype A1061 complexed with oligosaccharide receptor. | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Coat protein VP1, Coat protein VP2, ... | | Authors: | Fry, E.E, Newman, J.W, Curry, S, Najjam, S, Jackson, T, Blakemore, W, Lea, S.M, Miller, L, Burman, A, King, A.M, Stuart, D.I. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Foot-and-mouth disease virus serotype A1061 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation.
J.Gen.Virol., 86, 2005
|
|
2LVN
 
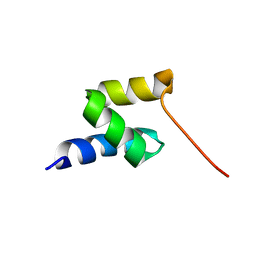 | | Structure of the gp78 CUE domain | | Descriptor: | E3 ubiquitin-protein ligase AMFR | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVO
 
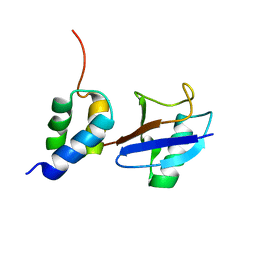 | | Structure of the gp78CUE domain bound to monubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVP
 
 | | gp78CUE domain bound to the distal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVQ
 
 | | gp78CUE domain bound to the proximal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LXP
 
 | | NMR structure of two domains in ubiquitin ligase gp78, RING and G2BR, bound to its conjugating enzyme Ube2g | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
2LXH
 
 | | NMR structure of the RING domain in ubiquitin ligase gp78 | | Descriptor: | E3 ubiquitin-protein ligase AMFR, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
1TAM
 
 | | HUMAN IMMUNODEFICIENCY VIRUS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 MATRIX PROTEIN | | Authors: | Matthews, S, Barlow, P, Clark, N, Kingsman, S, Kingsman, A, Campbell, I. | | Deposit date: | 1996-02-07 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of p17, the HIV matrix protein.
Biochem.Soc.Trans., 23, 1995
|
|
1TZ4
 
 | | [hPP19-23]-pNPY bound to DPC Micelles | | Descriptor: | neuropeptide Y,Pancreatic prohormone,neuropeptide Y | | Authors: | Lerch, M, Kamimori, H, Folkers, G, Aguilar, M.I, Beck-Sickinger, A.G, Zerbe, O. | | Deposit date: | 2004-07-09 | | Release date: | 2005-07-05 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Strongly Altered Receptor Binding Properties in PP and NPY Chimeras Are Accompanied by Changes
in Structure and Membrane Binding
Biochemistry, 44, 2005
|
|
1TZ5
 
 | | [pNPY19-23]-hPP bound to DPC Micelles | | Descriptor: | Pancreatic prohormone,neuropeptide Y,Pancreatic prohormone | | Authors: | Lerch, M, Kamimori, H, Folkers, G, Aguilar, M.I, Beck-Sickinger, A.G, Zerbe, O. | | Deposit date: | 2004-07-09 | | Release date: | 2005-07-05 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Strongly Altered Receptor Binding Properties in PP and NPY Chimeras Are Accompanied by Changes
in Structure and Membrane Binding
Biochemistry, 44, 2005
|
|
