2ADV
 
 | | Crystal Structures Of Glutaryl 7-Aminocephalosporanic Acid Acylase: mutational study of activation mechanism | | Descriptor: | Glutaryl 7- Aminocephalosporanic Acid Acylase | | Authors: | Kim, J.K, Yang, I.S, Shin, H.J, Cho, K.J, Ryu, E.K, Kim, S.H, Park, S.S, Kim, K.H. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Insight into autoproteolytic activation from the structure of cephalosporin acylase: a protein with two proteolytic chemistries.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2AE4
 
 | | Glutaryl 7-Aminocephalosporanic Acid Acylase: mutational study of activation mechanism | | Descriptor: | GLYCEROL, Glutaryl 7-Aminocephalosporanic Acid Acylase, SULFATE ION | | Authors: | Kim, J.K, Yang, I.S, Shin, H.J, Cho, K.J, Ryu, E.K, Kim, S.H, Park, S.S, Kim, K.H. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight into autoproteolytic activation from the structure of cephalosporin acylase: a protein with two proteolytic chemistries.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2AE3
 
 | | Glutaryl 7-Aminocephalosporanic Acid Acylase: mutational study of activation mechanism | | Descriptor: | GLYCEROL, Glutaryl 7-Aminocephalosporanic Acid Acylase | | Authors: | Kim, J.K, Yang, I.S, Shin, H.J, Cho, K.J, Ryu, E.K, Kim, S.H, Park, S.S, Kim, K.H. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into autoproteolytic activation from the structure of cephalosporin acylase: a protein with two proteolytic chemistries.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
5WE0
 
 | | Structural Basis for Shelterin Bridge Assembly | | Descriptor: | DNA-binding protein rap1, Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ... | | Authors: | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
5WE2
 
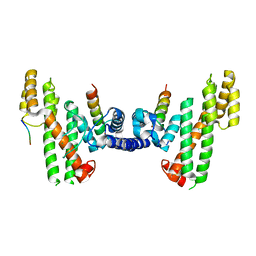 | | Structural Basis for Telomere Length Regulation by the Shelterin Bridge | | Descriptor: | DNA-binding protein rap1, Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ... | | Authors: | Kim, J.-K, Liu, J, Hu, X, Sankaran, B, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
8K2Y
 
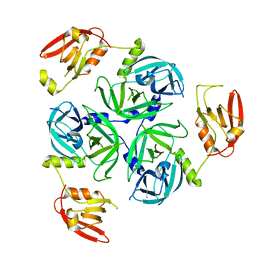 | | Crystal structure of MucD | | Descriptor: | serine endoprotease DegP-like protein MucD | | Authors: | Kim, J.H, Park, H.H. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of MucD from Pseudomonas syringae revealed N-terminal loop-mediated trimerization of HtrA-like serine protease.
Biochem.Biophys.Res.Commun., 688, 2023
|
|
1QNE
 
 | |
8YS9
 
 | |
4NAV
 
 | | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608 | | Descriptor: | HYPOTHETICAL PROTEIN XCC279 | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608
TO BE PUBLISHED
|
|
8JQV
 
 | |
6UKJ
 
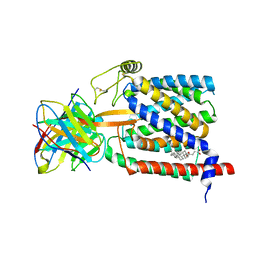 | | Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Chloroquine resistance transporter, Fab Heavy Chain, ... | | Authors: | Kim, J, Tan, Y.Z, Wicht, K.J, Erramilli, S.K, Dhingra, S.K, Okombo, J, Vendome, J, Hagenah, L.M, Giacometti, S.I, Warren, A.L, Nosol, K, Roepe, P.D, Potter, C.S, Carragher, B, Kossiakoff, A.A, Quick, M, Fidock, D.A, Mancia, F. | | Deposit date: | 2019-10-05 | | Release date: | 2019-12-04 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and drug resistance of the Plasmodium falciparum transporter PfCRT.
Nature, 576, 2019
|
|
8K6X
 
 | | Crystal structure of E.coli Cyanase complex with cyanate and bicarbonate | | Descriptor: | CARBONATE ION, Cyanate hydratase, SULFATE ION, ... | | Authors: | Kim, J, Nam, K.H, Cho, Y. | | Deposit date: | 2023-07-25 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural mechanism of Escherichia coli cyanase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8K6U
 
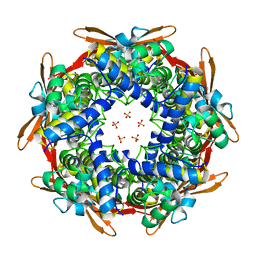 | |
8K6G
 
 | | Crystal structure of E.coli Cyanase | | Descriptor: | Cyanate hydratase, SULFATE ION | | Authors: | Kim, J, Nam, K.H, Cho, Y. | | Deposit date: | 2023-07-25 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural mechanism of Escherichia coli cyanase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8K6H
 
 | |
8K6S
 
 | |
7XCB
 
 | |
7XHY
 
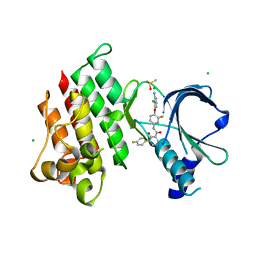 | | Crystal structure of MerTK Kinase domain with BMS794833 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Tyrosine-protein kinase Mer, ... | | Authors: | Kim, J.H, Lee, B.I. | | Deposit date: | 2022-04-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | BMS794833 inhibits macrophage efferocytosis by directly binding to MERTK and inhibiting its activity.
Exp.Mol.Med., 54, 2022
|
|
5X1E
 
 | |
6L5H
 
 | |
6L5J
 
 | |
7MBJ
 
 | |
4XB0
 
 | | Structure of the Plk2 polo-box domain | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Serine/threonine-protein kinase PLK2 | | Authors: | Kim, J.H, Ku, B, Kim, S.J. | | Deposit date: | 2014-12-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural analysis of the polo-box domain of human Polo-like kinase 2
Proteins, 83, 2015
|
|
8D73
 
 | | Crystal Structure of EGFR LRTM with compound 7 | | Descriptor: | (3S,4R)-3-fluoro-1-(4-{[4-(methylamino)-1-(propan-2-yl)pyrido[3,4-d]pyridazin-7-yl]amino}pyrimidin-2-yl)piperidin-4-ol, Epidermal growth factor receptor, GLYCEROL | | Authors: | Kim, J.L. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery of BLU-945, a Reversible, Potent, and Wild-Type-Sparing Next-Generation EGFR Mutant Inhibitor for Treatment-Resistant Non-Small-Cell Lung Cancer.
J.Med.Chem., 65, 2022
|
|
8D76
 
 | | Crystal Structure of EGFR LRTM with compound 24 | | Descriptor: | (3S,4R)-3-fluoro-1-(4-{[8-{3-[(methanesulfonyl)methyl]azetidin-1-yl}-5-(propan-2-yl)-2,7-naphthyridin-3-yl]amino}pyrimidin-2-yl)-3-methylpiperidin-4-ol, Epidermal growth factor receptor, GLYCEROL | | Authors: | Kim, J.L. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of BLU-945, a Reversible, Potent, and Wild-Type-Sparing Next-Generation EGFR Mutant Inhibitor for Treatment-Resistant Non-Small-Cell Lung Cancer.
J.Med.Chem., 65, 2022
|
|
