3PJF
 
 | |
3PJD
 
 | |
3PJE
 
 | |
4DIT
 
 | |
5GMN
 
 | | Crystal structure of human carbonic anhydrase II in complex with polmacoxib | | Descriptor: | 4-[3-(3-fluorophenyl)-5,5-dimethyl-4-oxidanylidene-furan-2-yl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into the inhibition of carbonic anhydrase by the COX-2-selective inhibitor polmacoxib (CG100649).
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GMM
 
 | | Crystal structure of human Carbonic anhydrase I in complex with polmacoxib | | Descriptor: | 4-[3-(3-fluorophenyl)-5,5-dimethyl-4-oxidanylidene-furan-2-yl]benzenesulfonamide, Carbonic anhydrase 1, ZINC ION | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural insight into the inhibition of carbonic anhydrase by the COX-2-selective inhibitor polmacoxib (CG100649).
Biochem. Biophys. Res. Commun., 478, 2016
|
|
3PG4
 
 | |
4KBZ
 
 | |
5X68
 
 | | Crystal Structure of Human KMO | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-21 | | Release date: | 2018-02-21 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5X6P
 
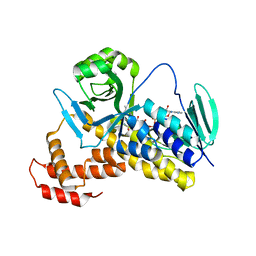 | | Crystal structure of Pseudomonas fluorescens KMO | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-22 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5YCS
 
 | |
5YCR
 
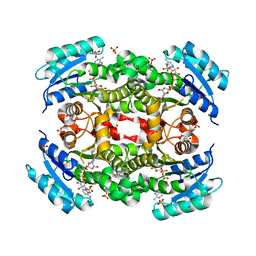 | |
5YCV
 
 | |
3V9B
 
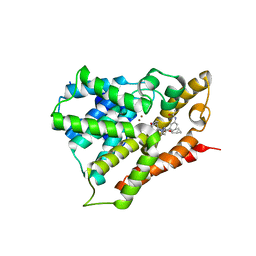 | |
5X6Q
 
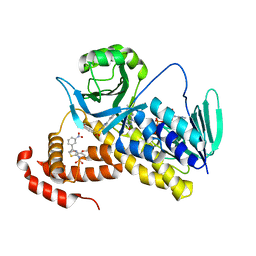 | | Crystal structure of Pseudomonas fluorescens KMO in complex with Ro 61-8048 | | Descriptor: | 3,4-dimethoxy-N-[4-(3-nitrophenyl)-1,3-thiazol-2-yl]benzenesulfonamide, FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-23 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5YCX
 
 | |
5X6R
 
 | |
5XY1
 
 | |
3JRX
 
 | | Crystal structure of the BC domain of ACC2 in complex with soraphen A | | Descriptor: | Acetyl-CoA carboxylase 2, SORAPHEN A | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
2HJW
 
 | | Crystal Structure of the BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2006-07-02 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the biotin carboxylase domain of human acetyl-CoA carboxylase 2.
Proteins, 70, 2008
|
|
3JRW
 
 | | Phosphorylated BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK
Biochem.Biophys.Res.Commun., 391, 2010
|
|
1ZC1
 
 | | Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites | | Descriptor: | Ubiquitin fusion degradation protein 1 | | Authors: | Park, S, Isaacson, R, Kim, H.T, Silver, P.A, Wagner, G. | | Deposit date: | 2005-04-10 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Ufd1 Exhibits the AAA-ATPase Fold with Two Distinct Ubiquitin Interaction Sites
Structure, 13, 2005
|
|
