5JH5
 
 | | Structural Basis for the Hierarchical Assembly of the Core of PRC1.1 | | Descriptor: | BCL-6 corepressor-like protein 1, Lysine-specific demethylase 2B, Polycomb group RING finger protein 1, ... | | Authors: | Wong, S.J, Taylor, A.B, Hart, P.J, Kim, C.A. | | Deposit date: | 2016-04-20 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | KDM2B Recruitment of the Polycomb Group Complex, PRC1.1, Requires Cooperation between PCGF1 and BCORL1.
Structure, 24, 2016
|
|
3IIA
 
 | | Crystal structure of apo (91-244) RIa subunit of cAMP-dependent protein kinase | | Descriptor: | GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Sjoberg, T.J, Kim, C, Kornev, A.P, Taylor, S.S. | | Deposit date: | 2009-07-31 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyclic AMP analog blocks kinase activation by stabilizing inactive conformation: conformational selection highlights a new concept in allosteric inhibitor design.
Mol.Cell Proteomics, 10, 2011
|
|
1SLH
 
 | | Mycobacterium tuberculosis dUTPase complexed with magnesium and dUDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-05 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SNF
 
 | | MYCOBACTERIUM TUBERCULOSIS DUTPASE COMPLEXED WITH MAGNESIUM AND DEOXYURIDINE 5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-10 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
3DPW
 
 | |
3DQO
 
 | |
6NAZ
 
 | | Crystal structure of DIRAS 2/3 chimera in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GTP-binding protein Di-Ras2,GTP-binding protein Di-Ras3,GTP-binding protein Di-Ras2, ... | | Authors: | Gilbert, Y.H, Reger, A, Sharma, R, Bast, J, Kim, C. | | Deposit date: | 2018-12-06 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.081 Å) | | Cite: | Crystal structure of DIRAS 2/3 chimera in complex with GDP
To Be Published
|
|
1SM8
 
 | | M. tuberculosis dUTPase complexed with chromium and dUTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHROMIUM ION, DEOXYURIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-08 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SJN
 
 | | Mycobacterium tuberculosis dUTPase complexed with magnesium and alpha,beta-imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-04 | | Release date: | 2004-03-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SMC
 
 | | Mycobacterium tuberculosis dUTPase complexed with dUTP in the absence of metal ion. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
5BV8
 
 | | G1324S mutation in von Willebrand Factor A1 domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, von Willebrand factor | | Authors: | Campbell, J.C, Kim, C, Tischer, A, Auton, M. | | Deposit date: | 2015-06-04 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Mutational Constraints on Local Unfolding Inhibit the Rheological Adaptation of von Willebrand Factor.
J.Biol.Chem., 291, 2016
|
|
3B4W
 
 | | Crystal structure of Mycobacterium tuberculosis aldehyde dehydrogenase complexed with NAD+ | | Descriptor: | Aldehyde dehydrogenase, ETHANOL, GLYCEROL, ... | | Authors: | Moon, J.H, Lyon, A.E, Yu, M, Hung, L.-W, Terwilliger, T, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-10-24 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of aldehyde dehydrogenase from Mycobacterium tuberculosis complexed with NAD+.
To Be Published
|
|
2K9J
 
 | | Integrin alphaIIb-beta3 transmembrane complex | | Descriptor: | Integrin alpha-IIb light chain, Integrin beta-3 | | Authors: | Lau, T, Kim, C, Ginsberg, M.H, Ulmer, T.S. | | Deposit date: | 2008-10-15 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling
Embo J., 28, 2009
|
|
1IF8
 
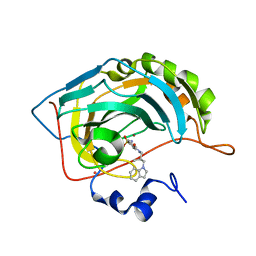 | | Carbonic Anhydrase II Complexed With (S)-N-(3-Indol-1-yl-2-methyl-propyl)-4-sulfamoyl-benzamide | | Descriptor: | (S)-N-(3-INDOL-1-YL-2-METHYL-PROPYL)-4-SULFAMOYL-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IF7
 
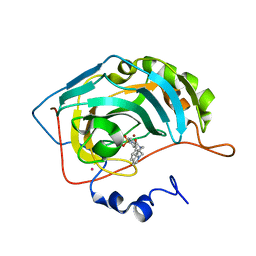 | | Carbonic Anhydrase II Complexed With (R)-N-(3-Indol-1-yl-2-methyl-propyl)-4-sulfamoyl-benzamide | | Descriptor: | (R)-N-(3-INDOL-1-YL-2-METHYL-PROPYL)-4-SULFAMOYL-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IF9
 
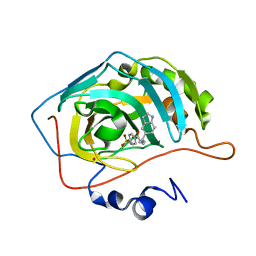 | | Carbonic Anhydrase II Complexed With N-[2-(1H-Indol-5-yl)-butyl]-4-sulfamoyl-benzamide | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-[2-(1H-INDOL-5-YL)-BUTYL]-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4HPL
 
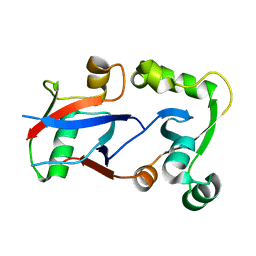 | | PCGF1 Ub fold (RAWUL)/BCOR PUFD Complex | | Descriptor: | BCL-6 corepressor, Polycomb group RING finger protein 1 | | Authors: | Junco, S.E, Wang, R, Gaipa, J, Taylor, A.B, Gearhart, M.D, Bardwell, V.J, Hart, P.J, Kim, C.A. | | Deposit date: | 2012-10-24 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Polycomb Group Protein PCGF1 in Complex with BCOR Reveals Basis for Binding Selectivity of PCGF Homologs.
Structure, 21, 2013
|
|
5JIZ
 
 | | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-pCPT-cGMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-amino-8-[(4-chlorophenyl)sulfanyl]-9-[(2S,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-3,9-dihydro-6H-purin-6-one, CALCIUM ION, ... | | Authors: | Campbell, J.C, Kim, C.W. | | Deposit date: | 2016-04-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-pCPT-cGMP
To Be Published
|
|
2QO3
 
 | | Crystal Structure of [KS3][AT3] didomain from module 3 of 6-deoxyerthronolide B synthase | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, ACETATE ION, ... | | Authors: | Khosla, C, Cane, E.D, Tang, Y, Chen, Y.A, Kim, C.Y. | | Deposit date: | 2007-07-19 | | Release date: | 2007-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase
Chem.Biol., 14, 2007
|
|
2FGG
 
 | | Crystal Structure of Rv2632c | | Descriptor: | Hypothetical protein Rv2632c/MT2708 | | Authors: | Yu, M, Bursey, E.H, Radhakannan, T, Segelke, B.W, Lekin, T, Toppani, D, Kim, C.Y, Kaviratne, T, Woodruff, T, Terwilliger, T.C, Hung, L.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-12-21 | | Release date: | 2006-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Rv2632c
To be Published
|
|
4HPM
 
 | | PCGF1 Ub fold (RAWUL)/BCORL1 PUFD Complex | | Descriptor: | BCL-6 corepressor-like protein 1, PHOSPHATE ION, Polycomb group RING finger protein 1 | | Authors: | Junco, S.E, Wang, R, Gaipa, J, Taylor, A.B, Gearhart, M.D, Bardwell, V.J, Hart, P.J, Kim, C.A. | | Deposit date: | 2012-10-24 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Polycomb Group Protein PCGF1 in Complex with BCOR Reveals Basis for Binding Selectivity of PCGF Homologs.
Structure, 21, 2013
|
|
3J1Z
 
 | | Inward-Facing Conformation of the Zinc Transporter YiiP revealed by Cryo-electron Microscopy | | Descriptor: | Cation efflux family protein | | Authors: | Coudray, N, Valvo, S, Hu, M, Lasala, R, Kim, C, Vink, M, Zhou, M, Provasi, D, Filizola, M, Tao, J, Fang, J, Penczek, P.A, Ubarretxena-Belandia, I, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | Deposit date: | 2012-07-24 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2A7Y
 
 | | Solution Structure of the Conserved Hypothetical Protein Rv2302 from the Bacterium Mycobacterium tuberculosis | | Descriptor: | Hypothetical protein Rv2302/MT2359 | | Authors: | Buchko, G.W, Kim, C.-Y, Terwilliger, T.C, Kennedy, M.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-06 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the conserved hypothetical protein Rv2302 from Mycobacterium tuberculosis.
J.Bacteriol., 188, 2006
|
|
3IM4
 
 | | Crystal structure of cAMP-dependent Protein Kinase A Regulatory Subunit I alpha in complex with dual-specific A-Kinase Anchoring Protein 2 | | Descriptor: | Dual specificity A kinase-anchoring protein 2, ZINC ION, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Sarma, G.N, Kinderman, F.S, Kim, C, von Daake, S, Taylor, S.S. | | Deposit date: | 2009-08-09 | | Release date: | 2010-02-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Structure of D-AKAP2:PKA RI Complex: Insights into AKAP Specificity and Selectivity
Structure, 18, 2010
|
|
7UBO
 
 | | Crystal Structure of the first bromodomain of human BRDT in complex with the inhibitor CCD-956 | | Descriptor: | Bromodomain testis-specific protein, DIMETHYL SULFOXIDE, N-[(2R)-1-(methylamino)-3-{1-[(4-methyl-2-oxo-1,2-dihydroquinolin-6-yl)acetyl]piperidin-4-yl}-1-oxopropan-2-yl]-5-phenylpyridine-2-carboxamide | | Authors: | Ta, H.M, Modukuri, R.K, Yu, Z, Tan, Z, Matzuk, M.M, Kim, C. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of potent BET bromodomain 1 stereoselective inhibitors using DNA-encoded chemical library selections.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
