6RR0
 
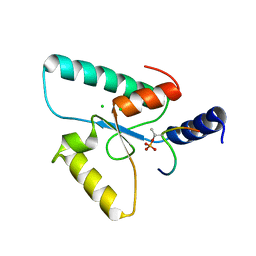 | | Crystal structure of the Sir4 H-BRCT domain in complex with Ubp10 pT123 peptide | | 分子名称: | CHLORIDE ION, Regulatory protein SIR4, Ubiquitin carboxyl-terminal hydrolase 10 | | 著者 | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | 登録日 | 2019-05-16 | | 公開日 | 2019-09-18 | | 最終更新日 | 2019-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
4UMG
 
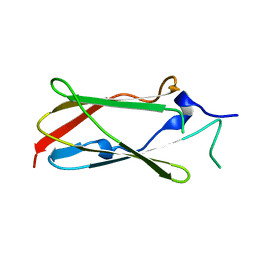 | | Crystal structure of the Lin-41 filamin domain | | 分子名称: | PROTEIN LIN-41 | | 著者 | Tocchini, C, Keusch, J.J, Miller, S.B, Finger, S, Gut, H, Stadler, M, Ciosk, R. | | 登録日 | 2014-05-16 | | 公開日 | 2014-10-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | The Trim-Nhl Protein Lin-41 Controls the Onset of Developmental Plasticity in Caenorhabditis Elegans.
Plos Genet., 10, 2014
|
|
6I3H
 
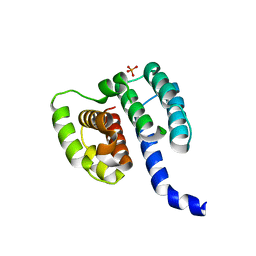 | | Crystal structure of influenza A virus M1 N-terminal domain (G18A mutation) | | 分子名称: | Matrix protein 1, PHOSPHATE ION | | 著者 | Miyake, Y, Keusch, J.J, Decamps, L, Ho-Xuan, H, Iketani, S, Gut, H, Kutay, U, Helenius, A, Yamauchi, Y. | | 登録日 | 2018-11-06 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Influenza virus uses transportin 1 for vRNP debundling during cell entry.
Nat Microbiol, 4, 2019
|
|
6QTM
 
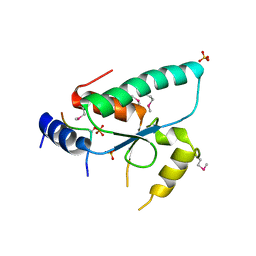 | | Crystal structure of the Sir4 H-BRCT domain in complex with Ty5 pS1095 peptide | | 分子名称: | Regulatory protein SIR4, Ribonuclease H, SULFATE ION | | 著者 | Gut, H, Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M. | | 登録日 | 2019-02-25 | | 公開日 | 2019-09-18 | | 最終更新日 | 2021-08-11 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
6QSZ
 
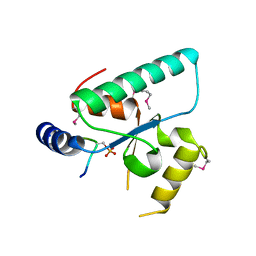 | | Crystal structure of the Sir4 H-BRCT domain in complex with Esc1 pS1450 peptide | | 分子名称: | CHLORIDE ION, Regulatory protein SIR4, Silent chromatin protein ESC1 | | 著者 | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | 登録日 | 2019-02-22 | | 公開日 | 2019-09-18 | | 最終更新日 | 2019-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
6FQ3
 
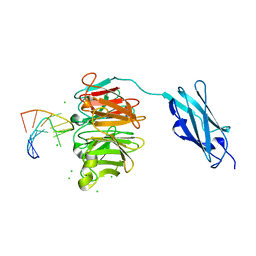 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with lin-29A 5'UTR 13mer RNA | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*GP*GP*AP*GP*UP*CP*CP*AP*AP*CP*UP*CP*C)-3') | | 著者 | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | 登録日 | 2018-02-13 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6FPT
 
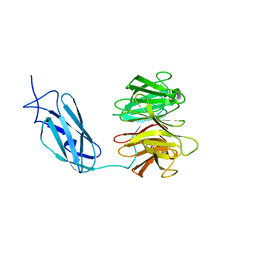 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains | | 分子名称: | E3 ubiquitin-protein ligase TRIM71 | | 著者 | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | 登録日 | 2018-02-12 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6FQL
 
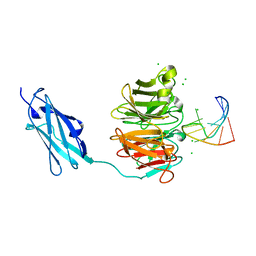 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with mab-10 3'UTR 13mer RNA | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*UP*GP*CP*AP*UP*UP*UP*AP*AP*UP*GP*CP*A)-3') | | 著者 | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | 登録日 | 2018-02-14 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.349 Å) | | 主引用文献 | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6RRV
 
 | | Crystal structure of the Sir4 H-BRCT domain | | 分子名称: | BROMIDE ION, CHLORIDE ION, Regulatory protein SIR4 | | 著者 | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | 登録日 | 2019-05-20 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
5G0F
 
 | | Crystal structure of Danio rerio HDAC6 ZnF-UBP domain | | 分子名称: | GLYCINE, HDAC6, NICKEL (II) ION, ... | | 著者 | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | 登録日 | 2016-03-18 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0H
 
 | | Crystal structure of Danio rerio HDAC6 CD2 in complex with (S)- trichostatin A | | 分子名称: | 1,2-ETHANEDIOL, HDAC6, POTASSIUM ION, ... | | 著者 | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | 登録日 | 2016-03-18 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0J
 
 | | Crystal structure of Danio rerio HDAC6 CD1 and CD2 (linker intact) in complex with Nexturastat A | | 分子名称: | CHLORIDE ION, HDAC6, NEXTURASTAT A, ... | | 著者 | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | 登録日 | 2016-03-18 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0I
 
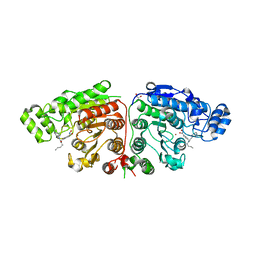 | | Crystal structure of Danio rerio HDAC6 CD1 and CD2 (linker cleaved) in complex with Nexturastat A | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, HDAC6, ... | | 著者 | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | 登録日 | 2016-03-18 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5OMB
 
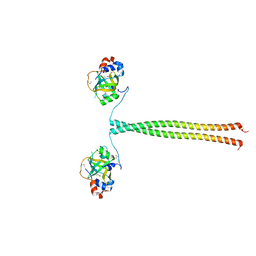 | | Crystal structure of K. lactis Ddc2 N-terminus in complex with S. cerevisiae Rfa1 N-OB domain | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA damage checkpoint protein LCD1, ... | | 著者 | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | 登録日 | 2017-07-28 | | 公開日 | 2017-10-25 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
5G0G
 
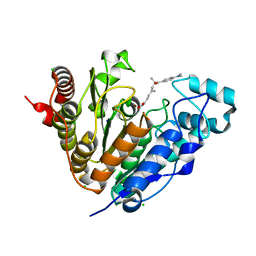 | | Crystal structure of Danio rerio HDAC6 CD1 in complex with trichostatin A | | 分子名称: | CHLORIDE ION, HDAC6, SODIUM ION, ... | | 著者 | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | 登録日 | 2016-03-18 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5OMC
 
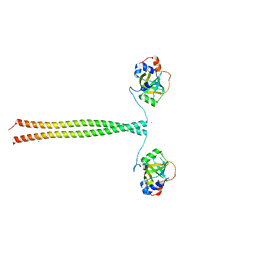 | | Crystal structure of K. lactis Ddc2 N-terminus in complex with S. cerevisiae Rfa1 (K45E mutant) N-OB domain | | 分子名称: | CHLORIDE ION, DNA damage checkpoint protein LCD1, Replication factor A protein 1 | | 著者 | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | 登録日 | 2017-07-28 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
5OMD
 
 | | Crystal structure of S. cerevisiae Ddc2 N-terminal coiled-coil domain | | 分子名称: | DNA damage checkpoint protein LCD1 | | 著者 | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | 登録日 | 2017-07-28 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
4AP6
 
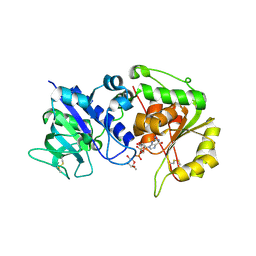 | | Crystal structure of human POFUT2 E54A mutant in complex with GDP- fucose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 2, ... | | 著者 | Chen, C, Keusch, J.J, Klein, D, Hess, D, Hofsteenge, J, Gut, H. | | 登録日 | 2012-03-30 | | 公開日 | 2012-08-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.401 Å) | | 主引用文献 | Structure of Human Pofut2: Insights Into Thrombospondin Type 1 Repeat Fold and O-Fucosylation.
Embo J., 31, 2012
|
|
4AE7
 
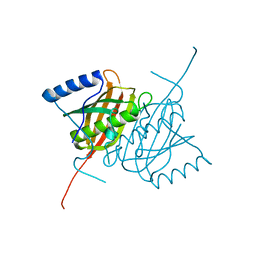 | | Crystal structure of human THEM5 | | 分子名称: | THIOESTERASE SUPERFAMILY MEMBER 5 | | 著者 | Zhuravleva, E, Gut, H, Hynx, D, Marcellin, D, Bleck, C.K.E, Genoud, C, Cron, P, Keusch, J.J, Dummler, B, Degli Esposti, M, Hemmings, B.A. | | 登録日 | 2012-01-09 | | 公開日 | 2012-07-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Acyl Coenzyme a Thioesterase Them5/Acot15 is Involved in Cardiolipin Remodeling and Fatty Liver Development.
Mol.Cell.Biol., 32, 2012
|
|
4AE8
 
 | | Crystal structure of human THEM4 | | 分子名称: | THIOESTERASE SUPERFAMILY MEMBER 4 | | 著者 | Zhuravleva, E, Gut, H, Hynx, D, Marcellin, D, Bleck, C.K.E, Genoud, C, Cron, P, Keusch, J.J, Dummler, B, Degli Esposti, M, Hemmings, B.A. | | 登録日 | 2012-01-09 | | 公開日 | 2012-07-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.594 Å) | | 主引用文献 | Acyl Coenzyme a Thioesterase Them5/Acot15 is Involved in Cardiolipin Remodeling and Fatty Liver Development.
Mol.Cell.Biol., 32, 2012
|
|
4AP5
 
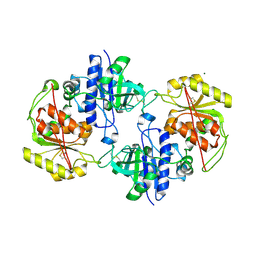 | | Crystal structure of human POFUT2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 2, ... | | 著者 | Chen, C, Keusch, J.J, Klein, D, Hess, D, Hofsteenge, J, Gut, H. | | 登録日 | 2012-03-30 | | 公開日 | 2012-08-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.003 Å) | | 主引用文献 | Structure of Human Pofut2: Insights Into Thrombospondin Type 1 Repeat Fold and O-Fucosylation.
Embo J., 31, 2012
|
|
3ZCO
 
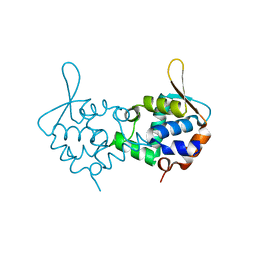 | | Crystal structure of S. cerevisiae Sir3 C-terminal domain | | 分子名称: | REGULATORY PROTEIN SIR3 | | 著者 | Oppikofer, M, Kueng, S, Keusch, J.J, Hassler, M, Ladurner, A.G, Gut, H, Gasser, S.M. | | 登録日 | 2012-11-21 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Dimerization of Sir3 Via its C-Terminal Winged Helix Domain is Essential for Yeast Heterochromatin Formation.
Embo J., 32, 2013
|
|
