2LRH
 
 | | Solution NMR structure of de novo designed protein, p-loop ntpase fold, Northeast Structural Genomics Consortium target or137 | | 分子名称: | De novo designed protein | | 著者 | Liu, G, Koga, N, Koga, R, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-03-30 | | 公開日 | 2012-07-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of de novo designed protein, p-loop ntpase fold, Northeast Structural Genomics Consortium target or137
To be Published
|
|
3NXF
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | 分子名称: | Retro-Aldolase, SULFATE ION | | 著者 | Althoff, E.A, Jiang, L, Wang, L, Lassila, J.K, Moody, J, Bolduc, J, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | 登録日 | 2010-07-13 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
3NNG
 
 | | Crystal structure of the F5/8 type C domain of Q5LFR2_BACFN protein from Bacteroides fragilis. Northeast Structural Genomics Consortium Target BfR258E | | 分子名称: | CALCIUM ION, uncharacterized protein | | 著者 | Vorobiev, S, Su, M, Dimaio, F, Baker, D, Seetharaman, J, Janjua, J, Xiao, R, Ciccosanti, C, Foote, E.L, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-06-23 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.177 Å) | | 主引用文献 | Crystal structure of the F5/8 type C domain of Q5LFR2_BACFN protein from Bacteroides fragilis.
To be Published
|
|
2LND
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, PFK fold, Northeast Structural Genomics Consortium Target OR134 | | 分子名称: | DE NOVO DESIGNED PROTEIN, PFK fold | | 著者 | Liu, G, Koga, N, Koga, R, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-12-23 | | 公開日 | 2012-01-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Northeast Structural Genomics Consortium Target OR134
To be Published
|
|
2LR0
 
 | | Solution NMR structure of de novo designed protein, p-loop ntpase fold, northeast structural genomics consortium target or136 | | 分子名称: | P-loop ntpase fold | | 著者 | Liu, G, Koga, N, Koga, R, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-03-19 | | 公開日 | 2012-07-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of de novo designed protein, p-loop ntpase fold, northeast structural genomics consortium target or136
To be Published
|
|
7CBC
 
 | |
4A91
 
 | | Crystal structure of the glutamyl-queuosine tRNAAsp synthetase from E. coli complexed with L-glutamate | | 分子名称: | GLUTAMIC ACID, GLUTAMYL-Q TRNA(ASP) SYNTHETASE, ZINC ION | | 著者 | Blaise, M, Olieric, V, Sauter, C, Lorber, B, Roy, B, Karmakar, S, Banerjee, R, Becker, H.D, Kern, D. | | 登録日 | 2011-11-22 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure of Glutamyl-Queuosine Trnaasp Synthetase Complexed with L-Glutamate: Structural Elements Mediating tRNA-Independent Activation of Glutamate and Glutamylation of Trnaasp Anticodon.
J.Mol.Biol., 381, 2008
|
|
2LVB
 
 | | Solution NMR Structure DE NOVO DESIGNED PFK fold PROTEIN, Northeast Structural Genomics Consortium (NESG) Target OR250 | | 分子名称: | DE NOVO DESIGNED PFK fold PROTEIN | | 著者 | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-06-30 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
4LED
 
 | | The Crystal Structure of Pyocin L1 bound to D-rhamnose at 2.37 Angstroms | | 分子名称: | Pyocin L1, alpha-D-rhamnopyranose | | 著者 | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, C.J, Walker, D. | | 登録日 | 2013-06-25 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
6B73
 
 | | Crystal Structure of a nanobody-stabilized active state of the kappa-opioid receptor | | 分子名称: | CHOLESTEROL, N-[(5alpha,6beta)-17-(cyclopropylmethyl)-3-hydroxy-7,8-didehydro-4,5-epoxymorphinan-6-yl]-3-iodobenzamide, Nanobody, ... | | 著者 | Che, T, Majumdar, S, Zaidi, S.A, Kormos, C, McCorvy, J.D, Wang, S, Mosier, P.D, Uprety, R, Vardy, E, Krumm, B.E, Han, G.W, Lee, M.Y, Pardon, E, Steyaert, J, Huang, X.P, Strachan, R.T, Tribo, A.R, Pasternak, G.W, Carroll, I.F, Stevens, R.C, Cherezov, V, Katritch, V, Wacker, D, Roth, B.L. | | 登録日 | 2017-10-03 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor.
Cell, 172, 2018
|
|
2LV8
 
 | | Solution NMR Structure de novo designed rossmann 2x2 fold protein, Northeast Structural Genomics Consortium (NESG) Target OR16 | | 分子名称: | De novo designed rossmann 2x2 fold protein | | 著者 | Liu, G, Koga, R, Koga, N, Xiao, R, Pederson, K, Hamilton, K, Ciccosanti, C, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-06-29 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
4LEA
 
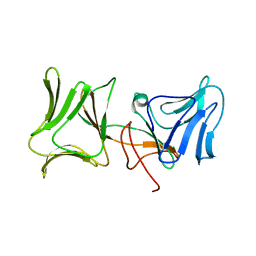 | | The Crystal Structure of Pyocin L1 bound to D-mannose at 2.55 Angstroms | | 分子名称: | Pyocin L1, beta-D-mannopyranose | | 著者 | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, C.J, Walker, D. | | 登録日 | 2013-06-25 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
4LE7
 
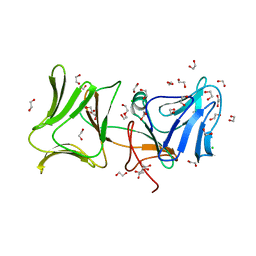 | | The Crystal Structure of Pyocin L1 at 2.09 Angstroms | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Pyocin L1 | | 著者 | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, R.J, Walker, D. | | 登録日 | 2013-06-25 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
2MBL
 
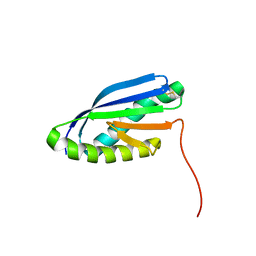 | | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33 | | 分子名称: | Top7 Fold Protein Top7m13 | | 著者 | Liu, G, Zanghellini, A.L, Chan, K, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-08-02 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33
To be Published
|
|
5TRV
 
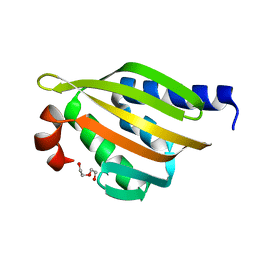 | | Crystal structure of a de novo designed protein with curved beta-sheet | | 分子名称: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | 著者 | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | 登録日 | 2016-10-27 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
2MR5
 
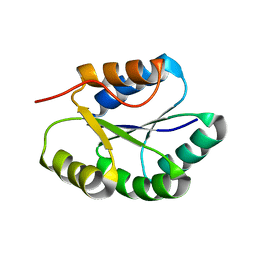 | | Solution NMR Structure of De novo designed Protein, Northeast Structural Genomics Consortium (NESG) Target OR457 | | 分子名称: | De novo designed Protein OR457 | | 著者 | Pulavarti, S, Nivon, L, Maglaqui, M, Janjua, H, Mao, L, Xiao, R, Kornhaber, G, Baker, D, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2014-07-01 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of De novo designed Protein, Northeast Structural Genomics Consortium (NESG) Target OR457
To be Published
|
|
2KL8
 
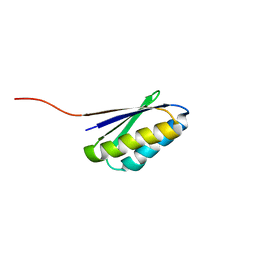 | | Solution NMR Structure of de novo designed ferredoxin-like fold protein, Northeast Structural Genomics Consortium Target OR15 | | 分子名称: | OR15 | | 著者 | Liu, G, Koga, N, Jiang, M, Koga, R, Xiao, R, Ciccosanti, C, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-06-30 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
3O6Y
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | 分子名称: | Retro-Aldolase, SULFATE ION | | 著者 | Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J.K, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | 登録日 | 2010-07-29 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.091 Å) | | 主引用文献 | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
5TPH
 
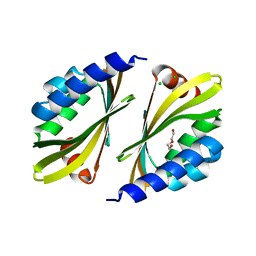 | | Crystal structure of a de novo designed protein homodimer with curved beta-sheet | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, de novo NTF2 homodimer | | 著者 | Basanta, B, Marcos, E, Oberdorfer, G, Chidyausiku, T.M, Sankaran, B, Baker, D. | | 登録日 | 2016-10-20 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5TS4
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | 分子名称: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | 著者 | Basanta, B, Oberdorfer, G, Chidyausiku, T.M, Marcos, E, Pereira, J.H, Sankaran, B, Zwart, P.H, Baker, D. | | 登録日 | 2016-10-27 | | 公開日 | 2017-01-25 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (3.005 Å) | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
2L82
 
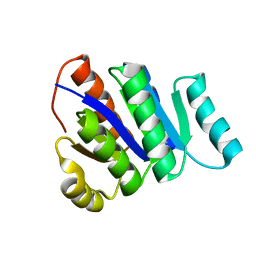 | | Solution NMR Structure of de novo designed protein, P-loop NTPase fold, Northeast Structural Genomics Consortium Target OR32 | | 分子名称: | DESIGNED PROTEIN OR32 | | 著者 | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Janjua, H, Tong, S, Acton, T.B, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-12-31 | | 公開日 | 2011-02-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Northeast Structural Genomics Consortium Target OR32
To be Published
|
|
6C83
 
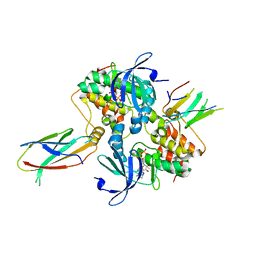 | | Structure of Aurora A (122-403) bound to inhibitory Monobody Mb2 and AMPPCP | | 分子名称: | Aurora kinase A, Mb2 Monobody, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | 著者 | Hoemberger, M, Kutter, S, Zorba, A, Nguyen, V, Shohei, A, Shohei, K, Kern, D. | | 登録日 | 2018-01-24 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Allosteric modulation of a human protein kinase with monobodies.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2LN3
 
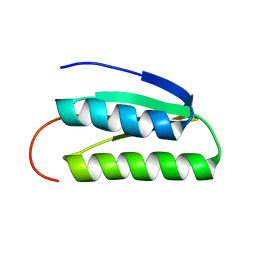 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, IF3-like fold, Northeast Structural Genomics Consortium Target OR135 (CASD target) | | 分子名称: | DE NOVO DESIGNED PROTEIN OR135 | | 著者 | Liu, G, Koga, R, Koga, N, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-12-15 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
2LTA
 
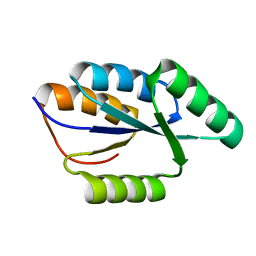 | | Solution NMR structure of De novo designed protein, rossmann 3x1 fold, Northeast Structural Genomics Consortium target OR157 | | 分子名称: | De novo designed protein | | 著者 | Liu, G, Koga, R, Koga, N, Xiao, R, Pederson, K, Hamilton, K, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-05-15 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
6BQG
 
 | | Crystal structure of 5-HT2C in complex with ergotamine | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, Ergotamine | | 著者 | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | 登録日 | 2017-11-27 | | 公開日 | 2018-02-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
