1XHY
 
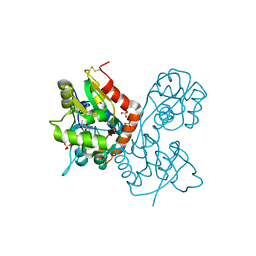 | | X-ray structure of the Y702F mutant of the GluR2 ligand-binding core (S1S2J) in complex with kainate at 1.85 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor, SULFATE ION | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-09-21 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
3HIZ
 
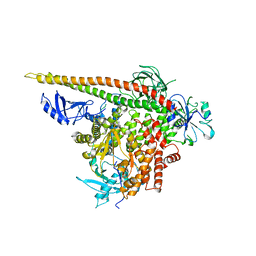 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | Deposit date: | 2009-05-20 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HHM
 
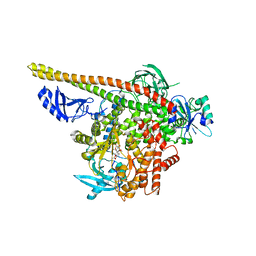 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha and the drug wortmannin | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | Deposit date: | 2009-05-15 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3RFN
 
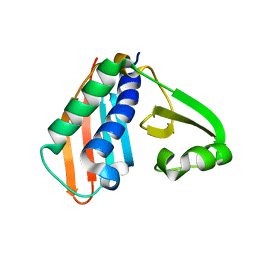 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | BB_1wnu_001, ZINC ION | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-06 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3RHU
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | SC_1wnu | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3RI0
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | BB_2cx5_001, GLYCEROL, SULFATE ION | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
2N0K
 
 | | Chemical shift assignments and structure of the alpha-crystallin domain from human, HSPB5 | | Descriptor: | Alpha-crystallin B chain | | Authors: | Rajagopal, P, Klevit, R.E, Shi, L, Baker, D. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis.
Elife, 4, 2015
|
|
2C8I
 
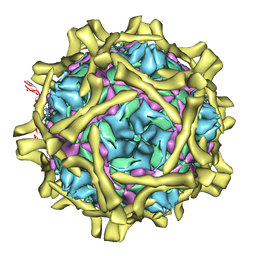 | | Complex Of Echovirus Type 12 With Domains 1, 2, 3 and 4 Of Its Receptor Decay Accelerating Factor (Cd55) By Cryo Electron Microscopy At 16 A | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, ECHOVIRUS 11 COAT PROTEIN VP1, ECHOVIRUS 11 COAT PROTEIN VP2, ... | | Authors: | Pettigrew, D.M, Williams, D.T, Kerrigan, D, Evans, D.J, Lea, S.M, Bhella, D. | | Deposit date: | 2005-12-05 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structural and Functional Insights Into the Interaction of Echoviruses and Decay-Accelerating Factor.
J.Biol.Chem., 281, 2006
|
|
3RIJ
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | GLYCEROL, SC_2cx5 | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-13 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
1OUR
 
 | | LecB (PA-LII) in complex with mannose | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
7FB8
 
 | | De Novo-Designed and Disulfide-Bridged Peptide Heterodimer - hd1 | | Descriptor: | ASP-ASP-LYS-ASP-CYS-ASP-GLU-TYR-CYS-LYS-LYS-THR-LYS-GLU-NH2, GLU-LE1-THR-GLY-HIS-ILE-GLU-GLY-PRO-THR-LE1-THR-LE1-HIS-CYS-LYS-NH2 | | Authors: | Yao, H, Yao, S, Zheng, Y, Moyer, A, Baker, D, Wu, C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | De novo design and directed folding of disulfide-bridged peptide heterodimers.
Nat Commun, 13, 2022
|
|
7FBA
 
 | | De Novo-Designed and Disulfide-Bridged Peptide Heterodimer - hd2 | | Descriptor: | ALA-LE1-CYS-GLU-CYS-GLY-PRO-THR-ARG-GLU-CYS-LYS-NH2, GLU-CYS-ARG-GLU-TYR-GLY-PRO-LE1-LYS-LE1-LE1-ALA-NH2 | | Authors: | Yao, H, Yao, S, Moyer, A, Zheng, Y, Baker, D, Wu, C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | De novo design and directed folding of disulfide-bridged peptide heterodimers.
Nat Commun, 13, 2022
|
|
3S64
 
 | | Saposin-like protein Ac-SLP-1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, Saposin-like protein 1 | | Authors: | Willis, C, Wang, C.K, Osman, A, Simon, A, Mulvenna, J, Pickering, D, Riboldi-Tunicliffe, A, Jones, M.K, Loukas, A, Hofmann, A. | | Deposit date: | 2011-05-25 | | Release date: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Insights into the membrane interactions of the saposin-like proteins Na-SLP-1 and Ac-SLP-1 from human and dog hookworm.
Plos One, 6, 2011
|
|
1OUX
 
 | | LecB (PA-LII) sugar-free | | Descriptor: | CALCIUM ION, SULFATE ION, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1OXC
 
 | | LecB (PA-LII) in complex with FUCOSE | | Descriptor: | CALCIUM ION, SULFATE ION, alpha-L-fucopyranose, ... | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-04-02 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
7JH5
 
 | | Co-LOCKR: de novo designed protein switch | | Descriptor: | Co-LOCKR: de novo designed protein switch | | Authors: | Bick, M.J, Lajoie, M.J, Boyken, S.E, Sankaran, B, Baker, D. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Designed protein logic to target cells with precise combinations of surface antigens.
Science, 369, 2020
|
|
3S63
 
 | | Saposin-like protein Na-SLP-1 | | Descriptor: | Saposin-like protein | | Authors: | Willis, C, Wang, C.K, Osman, A, Simon, A, Mulvenna, J, Pickering, D, Riboldi-Tunicliffe, A, Jones, M.K, Loukas, A, Hofmann, A. | | Deposit date: | 2011-05-24 | | Release date: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the membrane interactions of the saposin-like proteins Na-SLP-1 and Ac-SLP-1 from human and dog hookworm.
Plos One, 6, 2011
|
|
1PUX
 
 | | NMR Solution Structure of BeF3-Activated Spo0F, 20 conformers | | Descriptor: | Sporulation initiation phosphotransferase F | | Authors: | Gardino, A.K, Volkman, B.F, Cho, H.S, Lee, S.Y, Wemmer, D.E, Kern, D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of BeF(3)(-)-activated Spo0F reveals the conformational switch in a phosphorelay system.
J.Mol.Biol., 331, 2003
|
|
7K3H
 
 | | Crystal structure of deep network hallucinated protein 0217 | | Descriptor: | Network hallucinated protein 0217 | | Authors: | Pellock, S.J, Anishchenko, I, Chidyausiku, T.M, Bera, A.K, DiMaio, F, Baker, D. | | Deposit date: | 2020-09-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo protein design by deep network hallucination.
Nature, 600, 2021
|
|
1OUS
 
 | | Lecb (PA-LII) calcium-free | | Descriptor: | SULFATE ION, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1OVS
 
 | | LecB (PA-LII) in complex with core trimannoside | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose, ... | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-27 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1OVP
 
 | | LecB (PA-LII) in complex with fructose | | Descriptor: | CALCIUM ION, beta-D-fructopyranose, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-27 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
3SR0
 
 | |
4C3R
 
 | | Structure of dephosphorylated Aurora A (122-403) bound to AMPPCP | | Descriptor: | AURORA KINASE A, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Zorba, A, Kutter, S, Cho, Y.-J, Kern, D. | | Deposit date: | 2013-08-26 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular Mechanism of Aurora a Kinase Autophosphorylation and its Allosteric Activation by Tpx2.
Elife, 3, 2014
|
|
4C3P
 
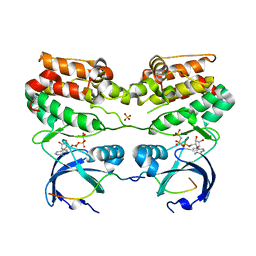 | | Structure of dephosphorylated Aurora A (122-403) bound to TPX2 and AMPPCP | | Descriptor: | AURORA KINASE A, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Zorba, A, Kutter, S, Kern, D. | | Deposit date: | 2013-08-26 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Molecular Mechanism of Aurora a Kinase Autophosphorylation and its Allosteric Activation by Tpx2.
Elife, 3, 2014
|
|
