3NWQ
 
 | |
1HYK
 
 | | AGOUTI-RELATED PROTEIN (87-132) (AC-AGRP(87-132)) | | Descriptor: | AGOUTI RELATED PROTEIN | | Authors: | Bolin, K.A, Anderson, D.J, Trulson, J.A, Thompson, D.A, Wilken, J, Kent, S.B.H, Millhauser, G.L. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a minimized human agouti related protein prepared by total chemical synthesis.
FEBS Lett., 451, 1999
|
|
5CY0
 
 | | Total Chemical Synthesis, Covalent Structure Verification, and X-ray Structure of Bioactive Ts3 Toxin by Racemic Protein Crystallography | | Descriptor: | GLYCEROL, Ts3 Toxin | | Authors: | Dang, B, Kubota, T, Mandal, K, Correa, A.M, Bezanilla, F, Kent, S.B. | | Deposit date: | 2015-07-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Elucidation of the Covalent and Tertiary Structures of Biologically Active Ts3 Toxin.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3E7R
 
 | | X-ray Crystal Structure of Racemic Plectasin | | Descriptor: | Plectasin | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-18 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods
Protein Sci., 18, 2009
|
|
3E7U
 
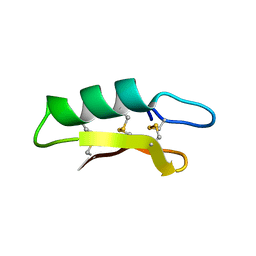 | | X-ray Crystal Structure of L-Plectasin | | Descriptor: | Plectasin | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-18 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods
Protein Sci., 18, 2009
|
|
3NXE
 
 | |
3NXN
 
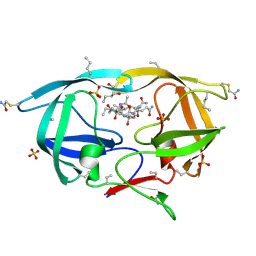 | | X-ray structure of ester chemical analogue 'covalent dimer' [Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, SULFATE ION, protease covalent dimer | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2010-07-14 | | Release date: | 2011-11-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NYG
 
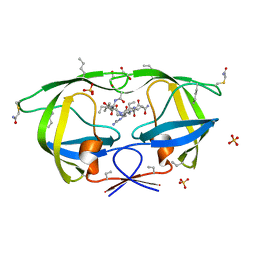 | |
2LET
 
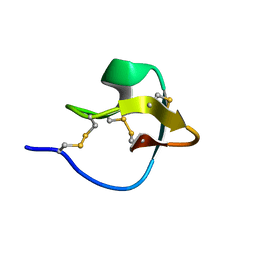 | | AN 1H NMR DETERMINATION OF THE THREE DIMENSIONAL STRUCTURES OF MIRROR IMAGE FORMS OF A LEU-5 VARIANT OF THE TRYPSIN INHIBITOR ECBALLIUM ELATERIUM (EETI-II) | | Descriptor: | TRYPSIN INHIBITOR II | | Authors: | Nielsen, K.J, Alewood, D, Andrews, J, Kent, S.B.H, Craik, D.J. | | Deposit date: | 1994-01-04 | | Release date: | 1994-05-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | An 1H NMR determination of the three-dimensional structures of mirror-image forms of a Leu-5 variant of the trypsin inhibitor from Ecballium elaterium (EETI-II).
Protein Sci., 3, 1994
|
|
3QTK
 
 | | The crystal structure of chemically synthesized VEGF-A | | Descriptor: | ACETATE ION, GLYCEROL, Vascular endothelial growth factor A, ... | | Authors: | Mandal, K, Kent, S.B.H. | | Deposit date: | 2011-02-22 | | Release date: | 2011-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Total chemical synthesis of biologically active vascular endothelial growth factor.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3E8Y
 
 | | Xray structure of scorpion toxin BmBKTx1 | | Descriptor: | CHLORIDE ION, Potassium channel toxin alpha-KTx 19.1, SULFATE ION | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-20 | | Release date: | 2009-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray structure of native scorpion toxin BmBKTx1 by racemic protein crystallography using direct methods.
J.Am.Chem.Soc., 131, 2009
|
|
5DL9
 
 | | Structure of Tetragonal Lysozyme in complex with Iodine solved by UWO Students | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IODIDE ION, ... | | Authors: | Bednarski, R, Cirricione, N, Greco, A, Hodgson, R, Kent, S, McGowan, J, Notherm, B, Patt, M, Vue, L, Bianchetti, C.M. | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of Tetragonal Lysozyme in complex with Iodine solved by UWO Students
To Be Published
|
|
5DLA
 
 | | Structure of Tetragonal Lysozyme solved by UWO Students | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Bednarski, R, Cirricione, N, Greco, A, Hodgson, R, Kent, S, McGowan, J, Notherm, B, Patt, M, Vue, L, Bianchetti, C.M. | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structure of Tetragonal Lysozyme in complex with Iodine solved by UWO Students
To Be Published
|
|
3NWX
 
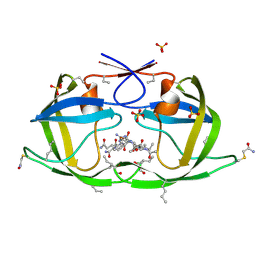 | | X-ray structure of ester chemical analogue [O-Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, SULFATE ION, protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2010-07-12 | | Release date: | 2011-11-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1B3A
 
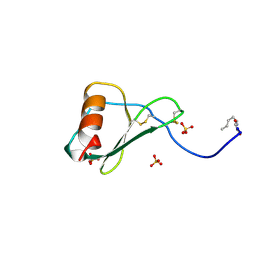 | | TOTAL CHEMICAL SYNTHESIS AND HIGH-RESOLUTION CRYSTAL STRUCTURE OF THE POTENT ANTI-HIV PROTEIN AOP-RANTES | | Descriptor: | PENTYLOXYAMINO-ACETALDEHYDE, PROTEIN (RANTES), SULFATE ION | | Authors: | Wilken, J, Hoover, D, Thompson, D.A, Barlow, P.N, Mcsparron, H, Picard, L, Wlodawer, A, Lubkowski, J, Kent, S.B.H. | | Deposit date: | 1998-12-07 | | Release date: | 1999-04-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Total chemical synthesis and high-resolution crystal structure of the potent anti-HIV protein AOP-RANTES.
Chem.Biol., 6, 1999
|
|
2NWD
 
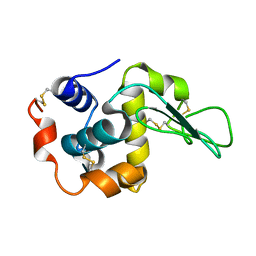 | |
4GLN
 
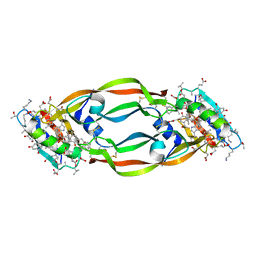 | | Crystal Structure of Chemically Synthesized Heterochiral {D-Protein Antagonist plus VEGF-A} Protein Complex in space group P21/n | | Descriptor: | D-RFX001, Vascular endothelial growth factor A | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2O40
 
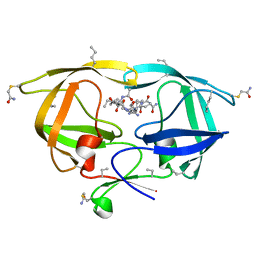 | |
4GLS
 
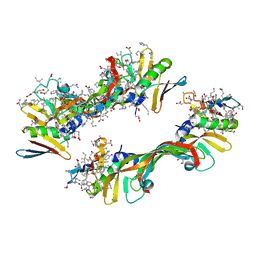 | | Crystal Structure of Chemically Synthesized Heterochiral {D-Protein Antagonist plus VEGF-A} Protein Complex in space group P21 | | Descriptor: | D- RFX001, D- Vascular endothelial growth factor-A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2NUI
 
 | | X-ray Structure of synthetic [D83A]RNase A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Boerema, D.J, Tereshko, V.A, Kent, S.B.H. | | Deposit date: | 2006-11-09 | | Release date: | 2007-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Total synthesis by modern chemical ligation methods and high resolution (1.1 A) X-ray structure of ribonuclease A
Biopolymers, 90, 2008
|
|
4GLU
 
 | | Crystal structure of the mirror image form of VEGF-A | | Descriptor: | ACETATE ION, D- Vascular endothelial growth factor-A, GLYCEROL, ... | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7HVP
 
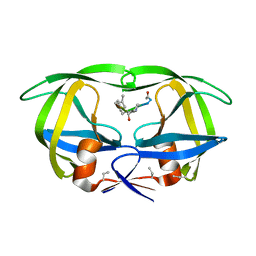 | | X-RAY CRYSTALLOGRAPHIC STRUCTURE OF A COMPLEX BETWEEN A SYNTHETIC PROTEASE OF HUMAN IMMUNODEFICIENCY VIRUS 1 AND A SUBSTRATE-BASED HYDROXYETHYLAMINE INHIBITOR | | Descriptor: | HIV-1 PROTEASE, INHIBITOR ACE-SER-LEU-ASN-PHE-PSI(CH(OH)-CH2N)-PRO-ILE VME (JG-365) | | Authors: | Swain, A.L, Miller, M.M, Green, J, Rich, D.H, Schneider, J, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1990-09-13 | | Release date: | 1993-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic structure of a complex between a synthetic protease of human immunodeficiency virus 1 and a substrate-based hydroxyethylamine inhibitor.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
3HDK
 
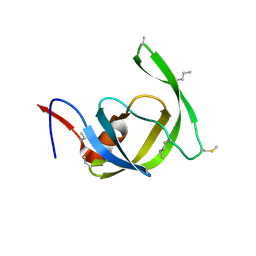 | | Crystal structure of chemically synthesized [Aib51/51']HIV-1 protease | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, [Aib51/51']HIV-1 protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2009-05-07 | | Release date: | 2010-04-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3HBO
 
 | | Crystal structure of chemically synthesized [D-Ala51/51']HIV-1 protease | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, [D-Ala51/51']HIV-1 protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2009-05-04 | | Release date: | 2010-05-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3DCK
 
 | | X-ray structure of D25N chemical analogue of HIV-1 protease complexed with ketomethylene isostere inhibitor | | Descriptor: | (2S)-2-{[(2R,5S)-5-{[(2S,3S)-2-{[(2S,3R)-2-(acetylamino)-3-hydroxybutanoyl]amino}-3-methylpentanoyl]amino}-2-butyl-4-oxononanoyl]amino}-N~1~-[(2S)-1-amino-5-carbamimidamido-1-oxopentan-2-yl]pentanediamide, Chemical analogue HIV-1 protease | | Authors: | Torbeev, V.Y, Mandal, K, Terechko, V.A, Kent, S.B.H. | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of chemically synthesized HIV-1 protease and a ketomethylene isostere inhibitor based on the p2/NC cleavage site
Bioorg.Med.Chem.Lett., 18, 2008
|
|
