4WFM
 
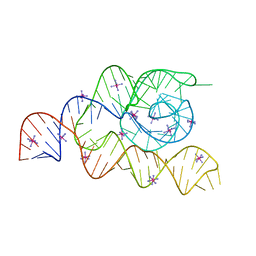 | | Structure of the complete bacterial SRP Alu domain | | 分子名称: | Bacillus subtilis small cytoplasmic RNA (scRNA),RNA, COBALT HEXAMMINE(III), MAGNESIUM ION | | 著者 | Kempf, G, Wild, K, Sinning, I. | | 登録日 | 2014-09-15 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of the complete bacterial SRP Alu domain.
Nucleic Acids Res., 42, 2014
|
|
4WFL
 
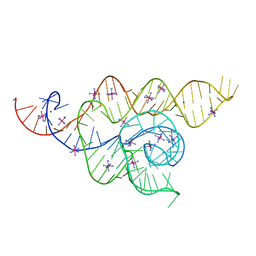 | |
7QNO
 
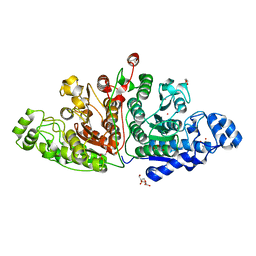 | | Crystal structure of ligand-free Danio rerio HDAC6 CD1 CD2 | | 分子名称: | GLYCEROL, Histone deacetylase 6, POTASSIUM ION, ... | | 著者 | Kempf, G, Langousis, G, Sanchez, J, Matthias, P. | | 登録日 | 2021-12-21 | | 公開日 | 2022-02-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Expression and Crystallization of HDAC6 Tandem Catalytic Domains.
Methods Mol.Biol., 2589, 2023
|
|
5NIY
 
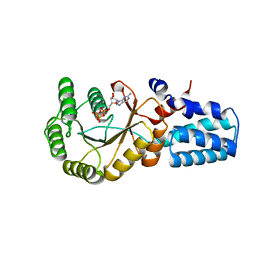 | | Signal recognition particle-docking protein FtsY | | 分子名称: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Signal recognition particle-docking protein FtsY | | 著者 | Kempf, G, Stjepanovic, G, Lapouge, K, Sinning, I. | | 登録日 | 2017-03-27 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Escherichia coli SRP Receptor Forms a Homodimer at the Membrane.
Structure, 26, 2018
|
|
2M7X
 
 | | Structural and Functional Analysis of Transmembrane Segment IV of the Salt Tolerance Protein Sod2 | | 分子名称: | Na(+)/H(+) antiporter | | 著者 | Ullah, A, Kemp, G, Lee, B, Alves, C, Young, H, Sykes, B.D, Fliegel, L. | | 登録日 | 2013-05-02 | | 公開日 | 2013-06-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Functional Analysis of Transmembrane Segment IV of the Salt Tolerance Protein Sod2.
J.Biol.Chem., 288, 2013
|
|
8PBA
 
 | |
6Y5E
 
 | | Structure of human cGAS (K394E) bound to the nucleosome (focused refinement of cGAS-NCP subcomplex) | | 分子名称: | Cyclic GMP-AMP synthase, DNA (153-MER), Histone H2A type 2-C, ... | | 著者 | Pathare, G.R, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2020-02-25 | | 公開日 | 2020-09-23 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structural mechanism of cGAS inhibition by the nucleosome.
Nature, 587, 2020
|
|
6Y5D
 
 | | Structure of human cGAS (K394E) bound to the nucleosome | | 分子名称: | Cyclic GMP-AMP synthase, DNA (153-MER), Histone H2A type 2-A, ... | | 著者 | Pathare, G.R, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2020-02-25 | | 公開日 | 2020-09-23 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural mechanism of cGAS inhibition by the nucleosome.
Nature, 587, 2020
|
|
6YOV
 
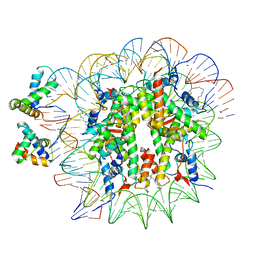 | | OCT4-SOX2-bound nucleosome - SHL+6 | | 分子名称: | DNA (142-MER), Green fluorescent protein,POU domain, class 5, ... | | 著者 | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2020-04-15 | | 公開日 | 2020-05-06 | | 最終更新日 | 2020-07-08 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
4UE5
 
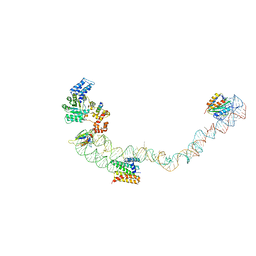 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | 分子名称: | 7S RNA, SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, ... | | 著者 | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | 登録日 | 2014-12-15 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UE4
 
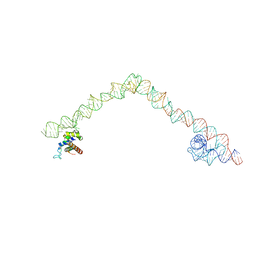 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | 分子名称: | 6S RNA, FTSQ SIGNAL SEQUENCE, SIGNAL RECOGNITION PARTICLE PROTEIN | | 著者 | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | 登録日 | 2014-12-15 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7Q3E
 
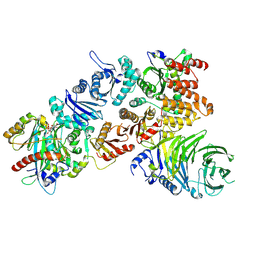 | | Structure of the mouse CPLANE-RSG1 complex | | 分子名称: | Ciliogenesis and planar polarity effector 2, GUANOSINE-5'-TRIPHOSPHATE, Protein fuzzy homolog, ... | | 著者 | Langousis, G, Cavadini, S, Kempf, G, Matthias, P. | | 登録日 | 2021-10-27 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structure of the ciliogenesis-associated CPLANE complex.
Sci Adv, 8, 2022
|
|
7Q3D
 
 | | Structure of the human CPLANE complex | | 分子名称: | Protein fuzzy homolog, Protein inturned, WD repeat-containing and planar cell polarity effector protein fritz homolog | | 著者 | Langousis, G, Cavadini, S, Kempf, G, Matthias, P. | | 登録日 | 2021-10-27 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structure of the ciliogenesis-associated CPLANE complex.
Sci Adv, 8, 2022
|
|
7OKQ
 
 | | Cryo-EM Structure of the DDB1-DCAF1-CUL4A-RBX1 Complex | | 分子名称: | Cullin-4A, DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, ... | | 著者 | Mohamed, W.I, Schenk, A.D, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2021-05-18 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (8.4 Å) | | 主引用文献 | The CRL4 DCAF1 cullin-RING ubiquitin ligase is activated following a switch in oligomerization state.
Embo J., 40, 2021
|
|
6T90
 
 | | OCT4-SOX2-bound nucleosome - SHL-6 | | 分子名称: | DNA (146-MER), Green fluorescent protein,POU domain, class 5, ... | | 著者 | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-10-25 | | 公開日 | 2020-05-06 | | 最終更新日 | 2020-07-08 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
6T93
 
 | | Nucleosome with OCT4-SOX2 motif at SHL-6 | | 分子名称: | DNA (153-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-10-25 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
8R1G
 
 | | Dimeric ternary structure of E6AP-E6-p53 | | 分子名称: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | 著者 | Sandate, C.R, Chakrabory, D, Kater, L, Kempf, G, Thoma, N.H. | | 登録日 | 2023-11-01 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | Structural insights into viral hijacking of p53 by E6 and E6AP
Biorxiv, 2023
|
|
8OSL
 
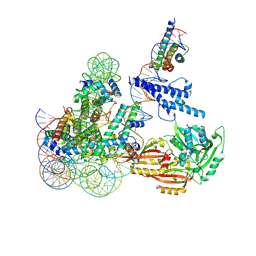 | | Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement) | | 分子名称: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (147-MER), ... | | 著者 | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | 登録日 | 2023-04-19 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSJ
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1) | | 分子名称: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | 著者 | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2023-04-19 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSK
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map) | | 分子名称: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | 著者 | Stoos, L, Michael, A.K, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2023-04-19 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OTS
 
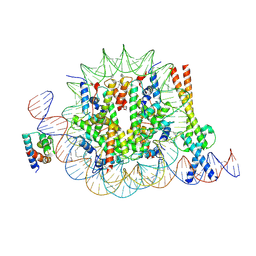 | | OCT4 and MYC-MAX co-bound to a nucleosome | | 分子名称: | DNA (127-MER), Green fluorescent protein,POU domain, class 5, ... | | 著者 | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | 登録日 | 2023-04-21 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OTT
 
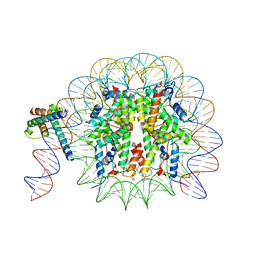 | | MYC-MAX bound to a nucleosome at SHL+5.8 | | 分子名称: | DNA (144-MER), Histone H2A type 1-B/E, Histone H2A type 1-K, ... | | 著者 | Stoos, L, Michael, A.K, Kempf, G, Kater, L, Cavadini, S, Thoma, N. | | 登録日 | 2023-04-21 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8R1F
 
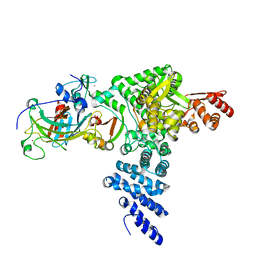 | | Monomeric E6AP-E6-p53 ternary complex | | 分子名称: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | 著者 | Sandate, C.R, Chakraborty, D, Kater, L, Kempf, G, Thoma, N.H. | | 登録日 | 2023-11-01 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Structural insights into viral hijacking of p53 by E6 and E6AP
Biorxiv, 2023
|
|
8P82
 
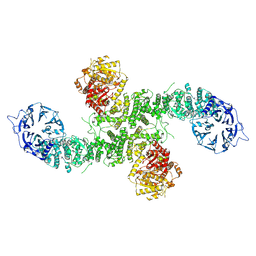 | | Cryo-EM structure of dimeric UBR5 | | 分子名称: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | 著者 | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2023-05-31 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
8BUM
 
 | | Structure of DDB1 bound to DS15-engaged CDK12-cyclin K | | 分子名称: | (2R)-2-[[6-(5-naphthalen-1-ylpentylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.36 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
