1YXL
 
 | | Crystal structure of a novel phospholipase A2 from Naja naja sagittifera at 1.5 A resolution | | Descriptor: | ACETIC ACID, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Singh, R.K, Jabeen, T, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2005-02-22 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | Crystal Structure of a novel phospholipase A2 from Naja naja sagittifera at 1.5 A resolution
To be Published
|
|
5WRF
 
 | | Crystal structure of dodecameric type II dehydroquinate dehydratase from Acinetobacter baumannii with unexplained connecting electron density between free cysteine residues of molecular pairs | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate dehydratase | | Authors: | Iqbal, N, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-01 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of dodecameric type II dehydroquinate dehydratase from Acinetobacter baumannii with unexplained connecting electron density between free cysteine residues of molecular pairs
To Be Published
|
|
1YF8
 
 | | Crystal structure of Himalayan mistletoe RIP reveals the presence of a natural inhibitor and a new functionally active sugar-binding site | | Descriptor: | 2-AMINO-4-ISOPROPYL-PTERIDINE-6-CARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mishra, V, Bilgrami, S, Sharma, R.S, Kaur, P, Yadav, S, Betzel, C, Babu, C.R, Singh, T.P. | | Deposit date: | 2004-12-31 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of himalayan mistletoe ribosome-inactivating protein reveals the presence of a natural inhibitor and a new functionally active sugar-binding site.
J.Biol.Chem., 280, 2005
|
|
5WUY
 
 | | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution | | Descriptor: | Chorismate synthase | | Authors: | Iqbal, N, Chaudhary, A, Shukla, K.P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution
To Be Published
|
|
1ZR8
 
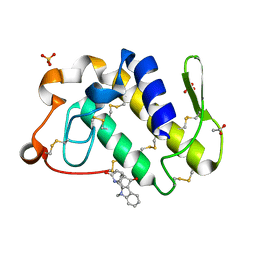 | | Crystal Structure of the complex formed between group II phospholipase A2 and a plant alkaloid ajmaline at 2.0A resolution | | Descriptor: | ACETIC ACID, AJMALINE, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Mahendra, M, Singh, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2005-05-19 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of the complex formed between group II phospholipase A2 and a plant alkaloid ajmaline at 2.0A resolution
To be Published
|
|
5X47
 
 | |
1ZWP
 
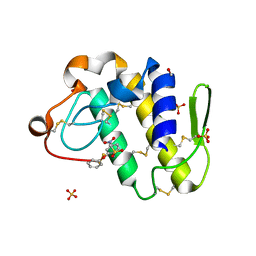 | | The atomic resolution Crystal structure of the Phospholipase A2 (PLA2) complex with Nimesulide reveals its weaker binding to PLA2 | | Descriptor: | 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2005-06-04 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The atomic resolution Crystal structure of the Phospholipase A2 (PLA2) complex with Nimesulide reveals its weaker binding to PLA2
To be Published
|
|
1P7V
 
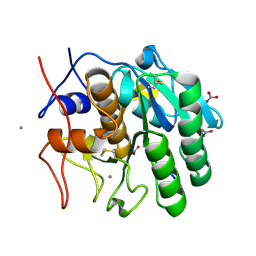 | | Structure of a complex formed between Proteinase K and a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ala-Ala at atomic resolution | | Descriptor: | CALCIUM ION, NITRATE ION, inhibitor peptide, ... | | Authors: | Bilgrami, S, Kaur, P, Chandra, V, Banumathi, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structure of a complex formed between Proteinase K and a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ala-Ala at atomic resolution
To be published
|
|
2B65
 
 | | Crystal structure of the complex of C-lobe of bovine lactoferrin with maltose at 1.5A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Singh, N, Prem kumar, R, Jabeen, T, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the complex of C-lobe of bovine lactoferrin with maltose at 1.5A resolution
To be published
|
|
5WV3
 
 | | Crystal structure of bovine lactoperoxidase with a partial Glu258-heme linkage at 2.07 A resolution. | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis of activation of mammalian heme peroxidases
Prog. Biophys. Mol. Biol., 133, 2018
|
|
2AYW
 
 | | Crystal Structure of the complex formed between trypsin and a designed synthetic highly potent inhibitor in the presence of benzamidine at 0.97 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-({[4-(DIAMINOMETHYL)PHENYL]AMINO}CARBONYL)-6-METHOXYPYRIDIN-3-YL]-5-{[(1-FORMYL-2,2-DIMETHYLPROPYL)AMINO]CARBONYL}BENZOIC ACID, BENZAMIDINE, ... | | Authors: | Sherawat, M, Kaur, P, Perbandt, M, Betzel, C, Slusarchyk, W.A, Bisacchi, G.S, Chang, C, Jacobson, B.L, Einspahr, H.M, Singh, T.P. | | Deposit date: | 2005-09-09 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure of the complex of trypsin with a highly potent synthetic inhibitor at 0.97 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2AOS
 
 | | Protein-protein Interactions of protective signalling factor: Crystal structure of ternary complex involving signalling protein from goat (SPG-40), tetrasaccharide and a tripeptide Trp-pro-Trp at 2.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Signaling protein from goat, SPG-40, ... | | Authors: | Kumar, J, Ethayathulla, A.S, Srivastava, D.B, Somvanshi, R.K, Singh, N, Sharma, S, Dey, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2005-08-14 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein-protein Interactions of protective signalling factor: Crystal structure of ternary complex involving signalling protein from goat (SPG-40), tetrasaccharide and a tripeptide Trp-pro-Trp at 2.9 A resolution
To be Published
|
|
1P7W
 
 | | Crystal structure of the complex of Proteinase K with a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ser-Ala at atomic resolution | | Descriptor: | CALCIUM ION, NITRATE ION, inhibitor peptide, ... | | Authors: | Bilgrami, S, Perbandt, M, Chandra, V, Banumathi, S, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the complex of Proteinase K with heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ser-Ala at atomic resolution
To be published
|
|
1OXR
 
 | | Aspirin induces its Anti-inflammatory effects through its specific binding to Phospholipase A2: Crystal structure of the complex formed between Phospholipase A2 and Aspirin at 1.9A resolution | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Singh, R.K, Ethayathulla, A.S, Jabeen, T, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2003-04-03 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Aspirin induces its anti-inflammatory effects through its specific binding to phospholipase A2: crystal structure of the complex formed between phospholipase A2 and aspirin at 1.9 angstroms resolution.
J.Drug Target., 13, 2005
|
|
1Y75
 
 | | A new form of catalytically inactive phospholipase A2 with an unusual disulphide bridge Cys 32- Cys 49 reveals recognition for N-acetylglucosmine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, phospholipase A2 isoform 5, ... | | Authors: | Jabeen, T, Singh, N, Jasti, J, Singh, R.K, Sharma, S, Perbandt, M, Betzel, C, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-12-08 | | Release date: | 2005-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a heterodimer of phospholipase A2 from Naja naja sagittifera at 2.3 A resolution reveals the presence of a new PLA2-like protein with a novel cys 32-Cys 49 disulphide bridge with a bound sugar at the substrate-binding site
Proteins, 62, 2006
|
|
1PO8
 
 | | Crystal structure of a complex formed between krait venom phospholipase A2 and heptanoic acid at 2.7 A resolution. | | Descriptor: | HEPTANOIC ACID, Phospholipase A2, SODIUM ION | | Authors: | Singh, G, Jayasankar, J, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2003-06-14 | | Release date: | 2004-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of a complex formed between krait venom phospholipase A2 and heptanoic acid at 2.7 A resolution.
To be Published
|
|
5Y48
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4-dione at 1.70 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, URACIL | | Authors: | Singh, P.K, Pandey, S, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
5YIH
 
 | | Crystal structure of tetrameric Nucleoside diphosphate kinase at 1.98 A resolution from Acinetobacter baumannii | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Bairagya, H.R, Sikarwar, J, Iqbal, N, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-10-04 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of tetrameric Nucleoside diphosphate kinase at 1.98 A resolution from Acinetobacter baumannii
To Be Published
|
|
5YPQ
 
 | |
5YHM
 
 | | Crystal structure of dehydroquinate dehydratase with tris induced oligomerisation at 1.907 Angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinate dehydratase, SULFATE ION | | Authors: | Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-09-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of dehydroquinate dehydratase with tris induced oligomerisation at 1.907 Angstrom resolution
To Be Published
|
|
1ZB5
 
 | | Recognition of peptide ligands by signalling protein from porcine mammary gland (SPP-40): Crystal structure of the complex of SPP-40 with a peptide Trp-Pro-Trp at 2.45A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PEPTIDE TRP-PRO-TRP, signal processing protein | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Singh, N, Somvanshi, R.K, Sharma, S, Dey, S, Kaur, P, Singh, T.P. | | Deposit date: | 2005-04-07 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Recognition of peptide ligands by signalling protein from porcine mammary gland (SPP-40): Crystal structure of the complex of SPP-40 with a peptide Trp-Pro-Trp at 2.45A resolution
To be Published
|
|
1OWS
 
 | | Crystal structure of a C49 Phospholipase A2 from Indian cobra reveals carbohydrate binding in the hydrophobic channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Phospholipase A2, ZINC ION | | Authors: | Jabeen, T, Jasti, J, Singh, N, Singh, R.K, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2003-03-30 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a C49 Phospholipase A2 from Indian cobra reveals carbohydrate binding in the hydrophobic channel
To be Published
|
|
1RMR
 
 | | Crystal Structure of Schistatin, a Disintegrin Homodimer from saw-scaled Viper (Echis carinatus) at 2.5 A resolution | | Descriptor: | Disintegrin schistatin | | Authors: | Bilgrami, S, Tomar, S, Yadav, S, Kaur, P, Kumar, J, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of schistatin, a disintegrin homodimer from saw-scaled viper (Echis carinatus) at 2.5 A resolution
J.Mol.Biol., 341, 2004
|
|
1ZBC
 
 | | Crystal Structure of the porcine signalling protein liganded with the peptide Trp-Pro-Trp (WPW) at 2.3 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3 mer peptide, signal processing protein | | Authors: | Srivastava, D.B, Kaur, P, Kumar, J, Somvanshi, R.K, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of the porcine signalling protein liganded with the peptide Trp-Pro-Trp (WPW) at 2.3 A resolution
To be Published
|
|
5YDB
 
 | | Crystal structure of the complex of type II dehydroquinate dehydratase from Acinetobacter baumannii with dehydroquinic acid at 1.76 Angstrom resolution | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase, SODIUM ION | | Authors: | Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of the complex of type II dehydroquinate dehydratase from Acinetobacter baumannii with dehydroquinic acid at 1.76 Angstrom resolution
To Be Published
|
|
