1WUK
 
 | | High resolution Structure Of The Oxidized State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
1WUL
 
 | | High Resolution Structure Of The Reduced State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
1WUI
 
 | | Ultra-High resolution Structure Of The Ni-A State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
7E6Z
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 50 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E6Y
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 1 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E6X
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 4 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E70
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 250 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E71
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 1 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7C86
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: Dark state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RETINAL, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-05-28 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7EK2
 
 | | Cryo-EM structure of VCCN1 in lipid nanodisc | | Descriptor: | Bestrophin-like protein | | Authors: | Hagino, T, Kato, T, Kasuya, G, Kobayashi, K, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of thylakoid-located voltage-dependent chloride channel VCCN1.
Nat Commun, 13, 2022
|
|
7EK1
 
 | | Cryo-EM structure of VCCN1 in detergent | | Descriptor: | Bestrophin-like protein | | Authors: | Hagino, T, Kato, T, Kasuya, G, Kobayashi, K, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of thylakoid-located voltage-dependent chloride channel VCCN1.
Nat Commun, 13, 2022
|
|
7EK3
 
 | | Cryo-EM structure of VCCN1 Y332A mutant in lipid nanodisc | | Descriptor: | Bestrophin-like protein | | Authors: | Hagino, T, Kato, T, Kasuya, G, Kobayashi, K, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of thylakoid-located voltage-dependent chloride channel VCCN1.
Nat Commun, 13, 2022
|
|
7CLR
 
 | | CryoEM structure of S.typhimurium flagellar LP ring | | Descriptor: | Flagellar L-ring protein, Flagellar P-ring protein | | Authors: | Yamaguchi, T, Makino, F, Miyata, T, Minamino, T, Kato, T, Namba, K. | | Deposit date: | 2020-07-21 | | Release date: | 2021-06-02 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the molecular bushing of the bacterial flagellar motor.
Nat Commun, 12, 2021
|
|
7D84
 
 | | 34-fold symmetry Salmonella S ring formed by full-length FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Kawamoto, A, Miyata, T, Makino, F, Kinoshita, M, Minamino, T, Imada, K, Kato, T, Namba, K. | | Deposit date: | 2020-10-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries.
Nat Commun, 12, 2021
|
|
7DZY
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 2490 | | Descriptor: | Fab Heavy chain of enhancing antibody 2490, Fab light chain of enhancing antibody 2490, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZX
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 8D2 | | Descriptor: | Fab Heavy chain of enhancing antibody, Fab light chain of enhancing antibody, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZW
 
 | | Apo spike protein from SARS-CoV2 | | Descriptor: | Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
3VXV
 
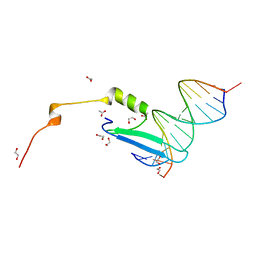 | | Crystal structure of methyl CpG Binding Domain of MBD4 in complex with the 5mCG/TG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*AP*CP*TP*AP*CP*(5CM)P*GP*GP*AP*CP*A)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-16 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
3VYB
 
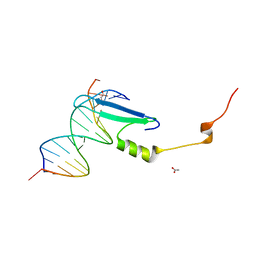 | | Crystal structure of methyl CpG binding domain of MBD4 in complex with the 5mCG/hmCG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*(5HC)P*GP*GP*TP*AP*GP*TP*GP*AP*CP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-22 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
3VXX
 
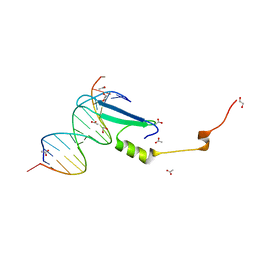 | | Crystal structure of methyl CpG binding domain of MBD4 in complex with the 5mCG/5mCG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*(5CM)P*GP*GP*TP*AP*GP*TP*GP*AP*CP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
3VYQ
 
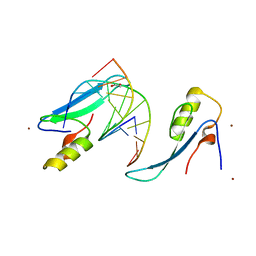 | | Crystal structure of the methyl CpG Binding Domain of MBD4 in complex with the 5mCG/TG sequence in space group P1 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*AP*TP*CP*(5CM)P*GP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*CP*TP*GP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
7WDT
 
 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with GlcNAc-6S | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ... | | Authors: | Yamada, C, Kashima, T, Fushinobu, S, Katoh, T, Katayama, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
7WDU
 
 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with PUGNAc-6S | | Descriptor: | Beta-N-acetylhexosaminidase, CALCIUM ION, [[(3R,4R,5S,6R)-3-acetamido-4,5-bis(oxidanyl)-6-(sulfooxymethyl)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Kashima, T, Yamada, C, Fushinobu, S, Katoh, T, Katayama, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
7Z4S
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with cyclic peptide GM4 including unnatural amino acids. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Owen, C.D, Miura, T, Malla, T, Lukacik, L, Strain-Damerell, C.M, Tumber, A, Brewitz, L, McDonough, M.A, Salah, E, Terasaka, N, Katoh, T, Kawamura, A, Schofield, C.J, Suga, H, Walsh, M.A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In vitro selection of macrocyclic peptide inhibitors containing cyclic gamma 2,4 -amino acids targeting the SARS-CoV-2 main protease.
Nat.Chem., 15, 2023
|
|
6L63
 
 | | Human Coagulation Factor XIIa (FXIIa) bound with the macrocyclic peptide F3 containing two (1S,2S)-2-ACHC residues | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL GROUP, ... | | Authors: | Sengoku, T, Katoh, T, Hirata, K, Suga, H, Ogata, K. | | Deposit date: | 2019-10-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ribosomal synthesis and de novo discovery of bioactive foldamer peptides containing cyclic beta-amino acids.
Nat.Chem., 12, 2020
|
|
