3WIP
 
 | | Crystal structure of acetylcholine bound to Ls-AChBP | | Descriptor: | ACETATE ION, ACETYLCHOLINE, Acetylcholine-binding protein, ... | | Authors: | Olsen, J.A, Balle, T, Gajhede, M, Ahring, P.K, Kastrup, J.S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition of the neurotransmitter acetylcholine by an acetylcholine binding protein reveals determinants of binding to nicotinic acetylcholine receptors
Plos One, 9, 2014
|
|
3U8L
 
 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3570 (1-(5-phenylpyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(5-phenylpyridin-3-yl)-1,4-diazepane, Acetylcholine-binding protein, SULFATE ION | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
1MQD
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-Des-Me-AMPA at 1.46 A resolution. Crystallization in the presence of lithium sulfate. | | Descriptor: | (S)-2-AMINO-3-(3-HYDROXY-ISOXAZOL-4-YL)PROPIONIC ACID, GLYCEROL, Glutamate receptor subunit 2, ... | | Authors: | Kasper, C, Lunn, M.-L, Liljefors, T, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2002-09-16 | | Release date: | 2003-07-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | GluR2 ligand-binding core complexes: Importance of the isoxazolol moiety and 5-substituent for the binding mode of AMPA-type agonists.
FEBS Lett., 531, 2002
|
|
4IGT
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the agonist ZA302 at 1.24A resolution | | Descriptor: | (4R)-4-{3-[hydroxy(methyl)amino]-3-oxopropyl}-L-glutamic acid, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Larsen, A.P, Venskutonyte, R, Gajhede, M, Kastrup, J.S, Frydenvang, K. | | Deposit date: | 2012-12-18 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Chemoenzymatic synthesis of new 2,4-syn-functionalized (S)-glutamate analogues and structure-activity relationship studies at ionotropic glutamate receptors and excitatory amino acid transporters.
J.Med.Chem., 56, 2013
|
|
4ISU
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (2R)-IKM-159 at 2.3A resolution. | | Descriptor: | (4aS,5aR,6R,8aS,8bS)-5a-(carboxymethyl)-8-oxo-2,4a,5a,6,7,8,8a,8b-octahydro-1H-pyrrolo[3',4':4,5]furo[3,2-b]pyridine-6-carboxylic acid, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Juknaite, L, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Studies on an (S)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid (AMPA) receptor antagonist IKM-159: asymmetric synthesis, neuroactivity, and structural characterization.
J.Med.Chem., 56, 2013
|
|
4KFQ
 
 | | Crystal structure of the NMDA receptor GluN1 ligand binding domain in complex with 1-thioxo-1,2-dihydro-[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one | | Descriptor: | 1-sulfanyl[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Steffensen, T.B, Tabrizi, F.M, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-04-27 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and pharmacological characterization of a novel N-methyl-D-aspartate (NMDA) receptor antagonist at the GluN1 glycine binding site.
J.Biol.Chem., 288, 2013
|
|
4NWD
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist (2S,4R)-4-(3-Methylamino-3-oxopropyl)glutamic acid at 2.6 A resolution | | Descriptor: | (4R)-4-[3-(methylamino)-3-oxopropyl]-L-glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Larsen, A.P, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
4NWC
 
 | | Crystal structure of the GluK3 ligand-binding domain (S1S2) in complex with the agonist (2S,4R)-4-(3-Methoxy-3-oxopropyl)glutamic acid at 2.01 A resolution. | | Descriptor: | (2S,4R)-4-(3-Methoxy-3-oxopropyl) glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Larsen, A.P, Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
2V3T
 
 | | Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in the apo form | | Descriptor: | CALCIUM ION, GLUTAMATE RECEPTOR DELTA-2 SUBUNIT SYNONYM GLURDELTA2, GLUR DELTA-2 | | Authors: | Naur, P, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-06-22 | | Release date: | 2007-08-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Ionotropic Glutamate-Like Receptor {Delta}2 Binds D-Serine and Glycine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2VAJ
 
 | | Crystal structure of NCAM2 Ig1 (I4122 cell unit) | | Descriptor: | NEURAL CELL ADHESION MOLECULE 2 | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Kastrup, J.S, Navarro-Poulsen, J.-C, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2007-08-31 | | Release date: | 2008-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of the Ig1 Domain of the Neural Cell Adhesion Molecule Ncam2 Displays Domain Swapping.
J.Mol.Biol., 382, 2008
|
|
2VRQ
 
 | | STRUCTURE OF AN INACTIVE MUTANT OF ARABINOFURANOSIDASE FROM THERMOBACILLUS XYLANILYTICUS IN COMPLEX WITH A PENTASACCHARIDE | | Descriptor: | ALPHA-L-ARABINOFURANOSIDASE, PHOSPHATE ION, alpha-L-arabinofuranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Paes, G, Skov, L.K, Odonohue, M.J, Remond, C, Kastrup, J.S, Gajhede, M, Mirza, O. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the Complex between a Branched Pentasaccharide and Thermobacillus Xylanilyticus Gh-51 Arabinofuranosidase Reveals Xylan-Binding Determinants and Induced Fit.
Biochemistry, 47, 2008
|
|
2VRK
 
 | | Structure of a seleno-methionyl derivative of wild type arabinofuranosidase from Thermobacillus xylanilyticus | | Descriptor: | ALPHA-L-ARABINOFURANOSIDASE, PHOSPHATE ION | | Authors: | Paes, G, Skov, L.K, ODonohue, M.J, Remond, C, Kastrup, J.S, Gajhede, M, Mirza, O. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Complex between a Branched Pentasaccharide and Thermobacillus Xylanilyticus Gh-51 Arabinofuranosidase Reveals Xylan-Binding Determinants and Induced Fit.
Biochemistry, 47, 2008
|
|
2V3U
 
 | | Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in complex with D-serine | | Descriptor: | CHLORIDE ION, D-SERINE, GLUTAMATE RECEPTOR DELTA-2 SUBUNIT, ... | | Authors: | Naur, P, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-06-22 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Ionotropic Glutamate-Like Receptor {Delta}2 Binds D-Serine and Glycine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2GDU
 
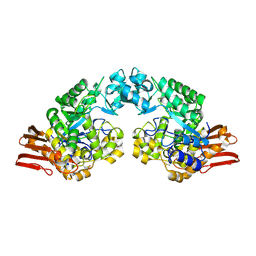 | | E232Q mutant of sucrose phosphorylase from BIFIDOBACTERIUM ADOLESCENTIS in complex with sucrose | | Descriptor: | beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, sucrose phosphorylase | | Authors: | Skov, L.K, Mirza, O, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2006-03-17 | | Release date: | 2006-09-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Rearrangements of Sucrose Phosphorylase from Bifidobacterium adolescentis during Sucrose Conversion
J.Biol.Chem., 281, 2006
|
|
3C9N
 
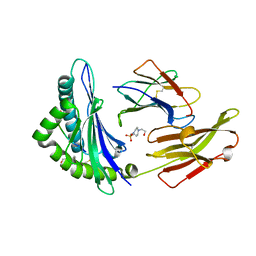 | | Crystal Structure of a SARS Corona Virus Derived Peptide Bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Roder, G.A, Kristensen, O, Kastrup, J.S, Buus, S, Gajhede, M. | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
3BFU
 
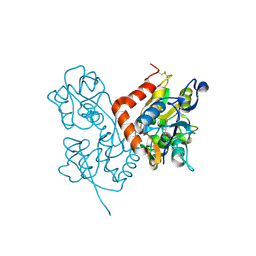 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (R)-TDPA at 1.95 A resolution | | Descriptor: | (2R)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, Glutamate receptor 2 | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
3FAS
 
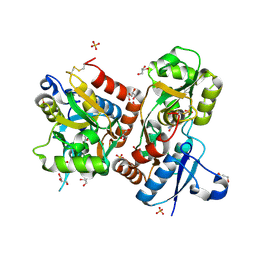 | | X-ray structure of iGluR4 flip ligand-binding core (S1S2) in complex with (S)-glutamate at 1.40A resolution | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 4, ... | | Authors: | Kasper, C, Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2008-11-18 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular mechanism of agonist recognition by the ligand-binding core of the ionotropic glutamate receptor 4
Febs Lett., 582, 2008
|
|
3FAT
 
 | | X-ray structure of iGluR4 flip ligand-binding core (S1S2) in complex with (S)-AMPA at 1.90A resolution | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, ACETIC ACID, GLYCEROL, ... | | Authors: | Kasper, C, Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2008-11-18 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism of agonist recognition by the ligand-binding core of the ionotropic glutamate receptor 4
Febs Lett., 582, 2008
|
|
1M5D
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH Br-HIBO AT 1.73 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5F
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH ACPA AT 1.95 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5C
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH Br-HIBO AT 1.65 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5E
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5B
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH 2-Me-Tet-AMPA AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-[3-HYDROXY-5-(2-METHYL-2H-TETRAZOL-5-YL)ISOXAZOL-4-YL]PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1PUP
 
 | |
4MH5
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate.
J.Struct.Biol., 176, 2011
|
|
