4QI9
 
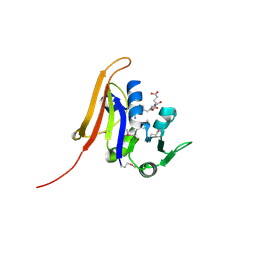 | | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate | | Descriptor: | Dihydrofolate reductase, METHOTREXATE | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate
To be Published
|
|
1QRH
 
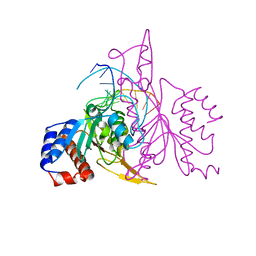 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE COMPLEXES WITH AN R145K MUTATION AT 2.7 A | | Descriptor: | 5'-(TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G*)-3', ECO RI ENDONCULEASE | | Authors: | Choi, J, Kim, Y, Greene, P, Hager, P, Rosenberg, J.M. | | Deposit date: | 1999-06-14 | | Release date: | 1999-06-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of the DNA-Eco RI Endonuclease Complexes with the ED144 and RK145 Mutations
To be Published
|
|
1QPS
 
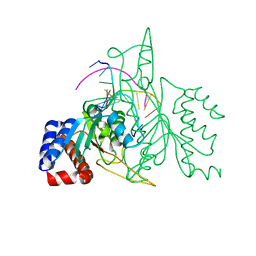 | | THE CRYSTAL STRUCTURE OF A POST-REACTIVE COGNATE DNA-ECO RI COMPLEX AT 2.50 A IN THE PRESENCE OF MN2+ ION | | Descriptor: | 5'-D(*AP*AP*TP*TP*CP*GP*CP*GP*)-3', 5'-D(*TP*CP*GP*CP*GP*)-3', ENDONUCLEASE ECORI, ... | | Authors: | Horvath, M, Choi, J, Kim, Y, Wilkosz, P, Rosenberg, J.M. | | Deposit date: | 1999-05-28 | | Release date: | 1999-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Integration of Recognition and Cleavage: X-Ray Structures of Pre- Transition State and Post-Reactive DNA-Eco RI Endonuclease Complexes
To be Published
|
|
5X90
 
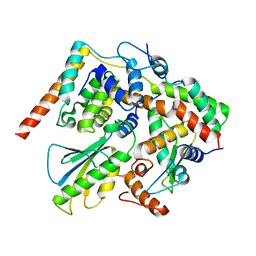 | | Structure of DotL(656-783)-IcmS-IcmW-LvgA derived from Legionella pneumophila | | Descriptor: | Hypothetical virulence protein, IcmO (DotL), IcmS, ... | | Authors: | Kim, H, Kwak, M.J, Kim, J.D, Kim, Y.G, Oh, B.H. | | Deposit date: | 2017-03-04 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
1QRI
 
 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE COMPLEXES WITH AN E144D MUTATION AT 2.7 A | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', ECO RI ENDONCULEASE | | Authors: | Choi, J, Kim, Y, Greene, P, Hager, P, Rosenberg, J.M. | | Deposit date: | 1999-06-14 | | Release date: | 1999-06-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure of the DNA-Eco RI Endonuclease Complexes with the ED144 and RK145 Mutations
To be Published
|
|
8D5I
 
 | | Crystal structure of dihydropteroate synthase H182G mutant (folP-SMZ_B27) from soil uncultured bacterium in complex with pteroic acid and pyrophosphate | | Descriptor: | PTEROIC ACID, PYROPHOSPHATE 2-, folP-SMZ_B27 | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Venkatesan, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of dihydropteroate synthase H182G mutant (folP-SMZ_B27) from soil uncultured bacterium in complex with pteroic acid
To Be Published
|
|
7LAP
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Xa | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, D(-)-TARTARIC ACID, ... | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-06 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
2MO0
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, TaCsp | | Descriptor: | Cold-shock DNA-binding domain protein | | Authors: | Jin, B, Jeong, K.W, Kim, Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and flexibility of the thermophilic cold-shock protein of Thermus aquaticus.
Biochem.Biophys.Res.Commun., 451, 2014
|
|
5X1U
 
 | |
1R0N
 
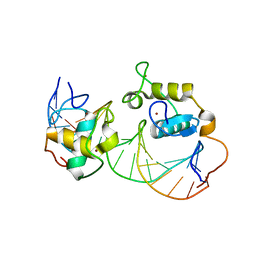 | | Crystal Structure of Heterodimeric Ecdsyone receptor DNA binding complex | | Descriptor: | Ecdsyone Response Element, Ecdysone Response Element, Ecdysone receptor, ... | | Authors: | Devarakonda, S, Harp, J.M, Kim, Y, Ozyhar, A, Rastinejad, F. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the heterodimeric Ecdysone Receptor DNA-binding complex
Embo J., 22, 2003
|
|
1R0O
 
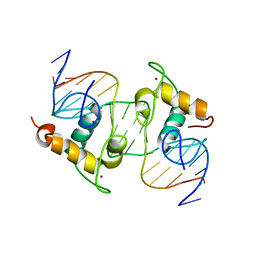 | | Crystal Structure of the Heterodimeric Ecdysone Receptor DNA-binding Complex | | Descriptor: | Ecdysone Response Element, Ecdysone receptor, Ultraspiracle protein, ... | | Authors: | Devarakonda, S, Harp, J.M, Kim, Y, Ozyhar, A, Rastinejad, F. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of the Heterodimeric Ecdysone Receptor DNA-binding Complex
Embo J., 22, 2003
|
|
1S6Y
 
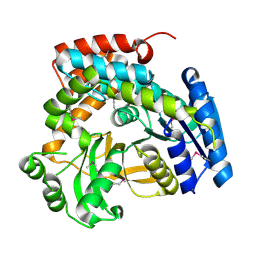 | | 2.3A crystal structure of phospho-beta-glucosidase | | Descriptor: | 6-phospho-beta-glucosidase | | Authors: | Tereshko, V, Dementieva, I, Kim, Y, Collat, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-28 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | 2.3A CRYSTAL STRUCTURE OF PHOSPHO-BETA-GLUCOSIDASE, licH Gene Product from BACILLUS STEAROTHERMOPHILUS
To be Published
|
|
1ROV
 
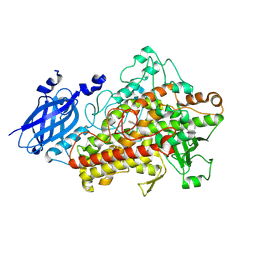 | | Lipoxygenase-3 Treated with Cumene Hydroperoxide | | Descriptor: | FE (III) ION, Seed lipoxygenase-3 | | Authors: | Vahedi-Faridi, A, Brault, P.A, Shah, P, Kim, Y.W, Dunham, W.R, Funk, M.O. | | Deposit date: | 2003-12-02 | | Release date: | 2004-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction between non-heme iron of lipoxygenases and cumene hydroperoxide: basis for enzyme activation, inactivation, and inhibition
J.Am.Chem.Soc., 126, 2004
|
|
1T8Q
 
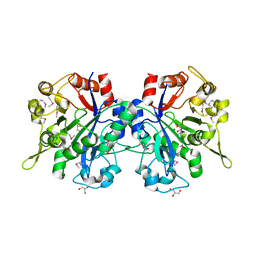 | | Structural genomics, Crystal structure of Glycerophosphoryl diester phosphodiesterase from E. coli | | Descriptor: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, periplasmic, ... | | Authors: | Zhang, R, Kim, Y, Dementieva, I, Duke, N, Stols, L, Donnelly, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Glycerophosphoryl diester phosphodiesterase from E. coli
To be Published
|
|
7E42
 
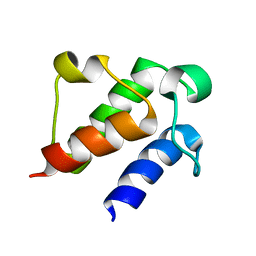 | |
7KAG
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, SULFATE ION | | Authors: | Stogios, P.J, Skarina, T, Chang, C, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-30 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2
To Be Published
|
|
5INT
 
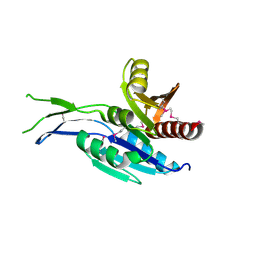 | | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC | | Descriptor: | Phosphopantothenate--cysteine ligase | | Authors: | Nocek, B, Zhou, M, Grimshaw, S, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-06 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC
To Be Published
|
|
5ZQE
 
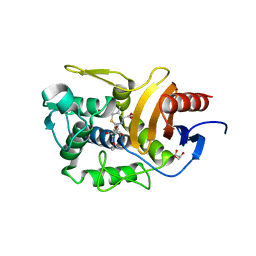 | | Crystal Structure of Penicillin-Binding Protein D2 from Listeria monocytogenes in the Cefuroxime bound form | | Descriptor: | 2-[CARBOXY-(2-FURAN-2-YL-2-METHOXYIMINO-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2018-04-18 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Crystal Structures of Penicillin-Binding Protein D2 from Listeria monocytogenes and Structural Basis for Antibiotic Specificity
Antimicrob. Agents Chemother., 62, 2018
|
|
2OCF
 
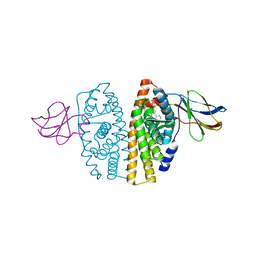 | | Human estrogen receptor alpha ligand-binding domain in complex with estradiol and the E2#23 FN3 monobody | | Descriptor: | ESTRADIOL, Estrogen receptor, Fibronectin | | Authors: | Rajan, S.S, Kuruvilla, S.M, Sharma, S.K, Kim, Y, Huang, J, Koide, A, Koide, S, Joachimiak, A, Greene, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5ZQD
 
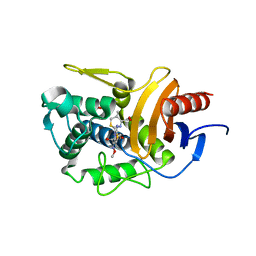 | |
5WKL
 
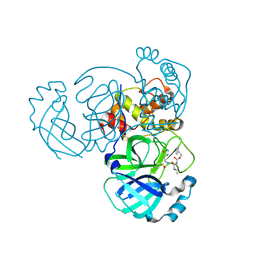 | | 1.85 A resolution structure of MERS 3CL protease in complex with piperidine-based peptidomimetic inhibitor 17 | | Descriptor: | (1R,2S)-2-{[N-({[4-benzyl-1-(tert-butoxycarbonyl)piperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[4-benzyl-1-(tert-butoxycarbonyl)piperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
1S9U
 
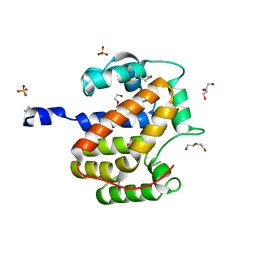 | | Atomic structure of a putative anaerobic dehydrogenase component | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, putative component of anaerobic dehydrogenases | | Authors: | Qiu, Y, Zhang, R, Tereshko, V, Kim, Y, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-05 | | Release date: | 2004-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The 1.38 A crystal structure of DmsD protein from Salmonella typhimurium, a proofreading chaperone on the Tat pathway.
Proteins, 71, 2008
|
|
5WKJ
 
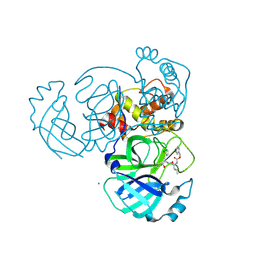 | | 2.05 A resolution structure of MERS 3CL protease in complex with inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, CALCIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
5WKK
 
 | | 1.55 A resolution structure of MERS 3CL protease in complex with inhibitor GC813 | | Descriptor: | (1R,2S)-2-[(N-{[2-(3-chlorophenyl)ethoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[2-(3-chlorophenyl)ethoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, MAGNESIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
5WKM
 
 | | 2.25 A resolution structure of MERS 3CL protease in complex with piperidine-based peptidomimetic inhibitor 21 | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)-4-ethylpiperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)-4-ethylpiperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
