2CDF
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5E) | | Descriptor: | TCR 5E | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
1GZQ
 
 | | CD1b in complex with Phophatidylinositol | | Descriptor: | 2-[(HYDROXY{[(2R,3R,5S,6R)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL]OXY}PHOSPHORYL)OXY]-1-[(PALMITOYLOXY)METHYL]ETHYL HEPTADECANOATE, B2-MICROGLOBULIN, DOCOSANE, ... | | Authors: | Gadola, S.D, Zaccai, N.R, Harlos, K, Shepherd, D, Ritter, G, Schmidt, R.R, Jones, E.Y, Cerundolo, V. | | Deposit date: | 2002-05-24 | | Release date: | 2002-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of Human Cd1B with Bound Ligands at 2.3 A, a Maze for Alkyl Chains
Nat.Immunol., 3, 2002
|
|
2CDE
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide specific T cell receptors - iNKT-TCR | | Descriptor: | INKT-TCR | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
1AKJ
 
 | | COMPLEX OF THE HUMAN MHC CLASS I GLYCOPROTEIN HLA-A2 AND THE T CELL CORECEPTOR CD8 | | Descriptor: | BETA 2-MICROGLOBULIN, HIV REVERSE TRANSCRIPTASE EPITOPE, MHC CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A*0201) (ALPHA CHAIN), ... | | Authors: | Tormo, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-05-21 | | Release date: | 1997-09-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the complex between human CD8alpha(alpha) and HLA-A2.
Nature, 387, 1997
|
|
1EPF
 
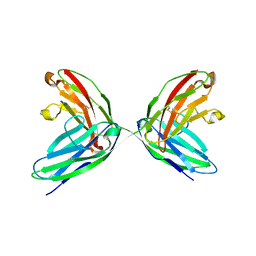 | | CRYSTAL STRUCTURE OF THE TWO N-TERMINAL IMMUNOGLOBULIN DOMAINS OF THE NEURAL CELL ADHESION MOLECULE (NCAM) | | Descriptor: | CALCIUM ION, PROTEIN (NEURAL CELL ADHESION MOLECULE) | | Authors: | Kasper, C, Rasmussen, H, Kastrup, J.S, Ikemizu, S, Jones, E.Y, Berezin, V, Bock, E, Larsen, I.K. | | Deposit date: | 2000-03-29 | | Release date: | 2000-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of cell-cell adhesion by NCAM.
Nat.Struct.Biol., 7, 2000
|
|
1A1M
 
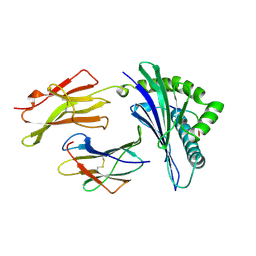 | | MHC CLASS I MOLECULE B*5301 COMPLEXED WITH PEPTIDE TPYDINQML FROM GAG PROTEIN OF HIV2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Harlos, K, Mcmichael, A.J, Stuart, D.I, Bell, J.I, Jones, E.Y. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53.
Immunity, 4, 1996
|
|
2CIK
 
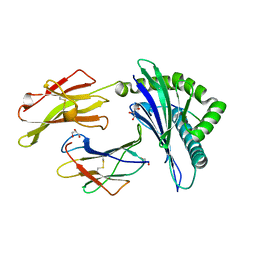 | | Insights Into Crossreactivity in Human Allorecognition: The Structure of HLA-B35011 Presenting an Epitope derived from Cytochrome P450. | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN B-35 ALPHA CHAIN, ... | | Authors: | Hourigan, C.S, Harkiolaki, M, Peterson, N.A, Bell, J.I, Jones, E.Y, O'Callaghan, C.A. | | Deposit date: | 2006-03-22 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structure of the Human Allo-Ligand Hla-B3501 in Complex with a Cytochrome P450 Peptide: Steric Hindrance Influences Tcr Allo-Recognition.
Eur.J.Immunol., 36, 2006
|
|
1BQU
 
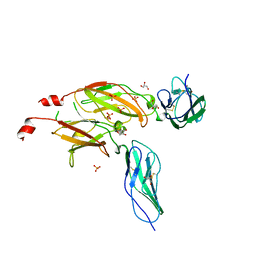 | | CYTOKYNE-BINDING REGION OF GP130 | | Descriptor: | GLYCEROL, PROTEIN (GP130), SULFATE ION | | Authors: | Bravo, J, Staunton, D, Heath, J.K, Jones, E.Y. | | Deposit date: | 1998-08-18 | | Release date: | 1998-08-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cytokine-binding region of gp130.
EMBO J., 17, 1998
|
|
1BND
 
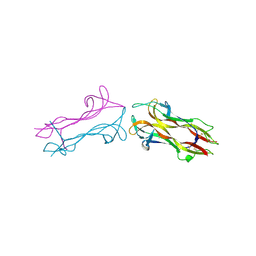 | | STRUCTURE OF THE BRAIN-DERIVED NEUROTROPHIC FACTOR(SLASH)NEUROTROPHIN 3 HETERODIMER | | Descriptor: | BRAIN DERIVED NEUROTROPHIC FACTOR, ISOPROPYL ALCOHOL, NEUROTROPHIN 3 | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1994-12-12 | | Release date: | 1996-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the brain-derived neurotrophic factor/neurotrophin 3 heterodimer.
Biochemistry, 34, 1995
|
|
2V2W
 
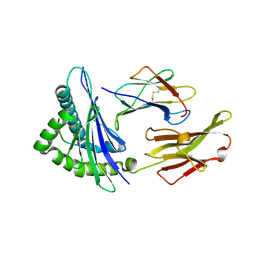 | | T CELL CROSS-REACTIVITY AND CONFORMATIONAL CHANGES DURING TCR ENGAGEMENT | | Descriptor: | BETA-2 MICROGLOBULIN, HIV P17, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Lee, J.K, Stewart-Jones, G, Dong, T, Harlos, K, Di Gleria, K, Dorrell, L, Douek, D.C, Van Der Merwe, P.A, Jones, E.Y, Mcmichael, A.J. | | Deposit date: | 2007-06-07 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | T Cell Cross-Reactivity and Conformational Changes During Tcr Engagement.
J.Exp.Med., 200, 2004
|
|
2V2X
 
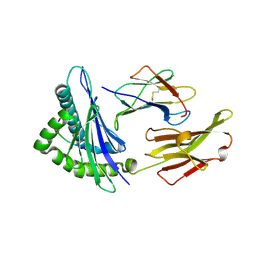 | | T cell cross-reactivity and conformational changes during TCR engagement. | | Descriptor: | BETA-2 MICROGLOBULIN, HIV P17, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Lee, J.K, Stewart-Jones, G, Dong, T, harlos, K, Di Gleria, K, Dorrell, L, Douek, D.C, van der Merwe, P.A, Jones, E.Y, McMichael, A.J. | | Deposit date: | 2007-06-07 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | T Cell Cross-Reactivity and Conformational Changes During Tcr Engagement.
J.Exp.Med., 200, 2004
|
|
6YV0
 
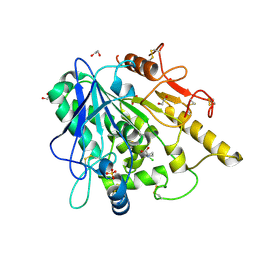 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLIDINE-3-CARBOXYLIC ACID FRAGMENT 587 | | Descriptor: | (3~{R})-1-(2-chlorophenyl)pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YUY
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 471 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methyl-5-(trifluoromethyl)-1~{H}-pyrrole-3-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Hillier, J, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YSK
 
 | | 1-phenylpyrroles and 1-enylpyrrolidines as inhibitors of Notum | | Descriptor: | (3~{S})-1-[4-chloranyl-3-(trifluoromethyl)phenyl]pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-04-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6ZYF
 
 | | Notum_Ghrelin complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-07-31 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Notum deacylates octanoylated ghrelin.
Mol Metab, 49, 2021
|
|
1OLZ
 
 | | The ligand-binding face of the semaphorins revealed by the high resolution crystal structure of SEMA4D | | Descriptor: | SEMAPHORIN 4D | | Authors: | Love, C.A, Harlos, K, Mavaddat, N, Davis, S.J, Stuart, D.I, Jones, E.Y, Esnouf, R.M. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ligand-Binding Face of the Semaphorins Revealed by the High-Resolution Crystal Structure of Sema4D
Nat.Struct.Biol., 10, 2003
|
|
1ODA
 
 | | N-terminal of Sialoadhesin in complex with Me-a-9-N-(biphenyl-4-carbonyl)-amino-9-deoxy-Neu5Ac (BIP compound) | | Descriptor: | ME-A-9-N-(BIPHENYL-4-CARBONYL)-AMINO-9-DEOXY-NEU5AC, SIALOADHESIN | | Authors: | Zaccai, N.R, Maenaka, K, Maenaka, T, Crocker, P.R, Brossmer, R, Kelm, S, Jones, E.Y. | | Deposit date: | 2003-02-14 | | Release date: | 2003-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure Guided Design of Sialic Acid-Based Siglec Inhibitors and Crystallographic Analysis in Complex with Sialoadhesin
Structure, 11, 2003
|
|
1OEB
 
 | | Mona/Gads SH3C domain | | Descriptor: | CADMIUM ION, GRB2-RELATED ADAPTOR PROTEIN 2, LYMPHOCYTE CYTOSOLIC PROTEIN 2 | | Authors: | Harkiolaki, M, Lewitzky, M, Gilbert, R.J.C, Jones, E.Y, Bourette, R.P, Mouchiroud, G, Sondermann, H, Moarefi, I, Feller, S.M. | | Deposit date: | 2003-03-24 | | Release date: | 2003-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for SH3 Domain-Mediated High-Affinity Binding between Mona/Gads and Slp-76
Embo J., 22, 2003
|
|
1QFO
 
 | | N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) IN COMPLEX WITH 3'SIALYLLACTOSE | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, PROTEIN (SIALOADHESIN) | | Authors: | May, A.P, Robinson, R.C, Vinson, M, Crocker, P.R, Jones, E.Y. | | Deposit date: | 1999-04-12 | | Release date: | 1999-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the N-terminal domain of sialoadhesin in complex with 3' sialyllactose at 1.85 A resolution.
Mol.Cell, 1, 1998
|
|
6ZUV
 
 | | Notum fragment 286 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 5-Phenyl-1,3,4-oxadiazol-2(3 H )-ones Are Potent Inhibitors of Notum Carboxylesterase Activity Identified by the Optimization of a Crystallographic Fragment Screening Hit.
J.Med.Chem., 63, 2020
|
|
1QQR
 
 | | CRYSTAL STRUCTURE OF STREPTOKINASE DOMAIN B | | Descriptor: | STREPTOKINASE DOMAIN B | | Authors: | Spraggon, G, Zhang, X.X, Ponting, C.P, Fox, V.F, Phillips, C, Smith, R.A.G, Jones, E.Y, Dobson, C, Stuart, D.I. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Streptokinse Domain B
To be Published
|
|
1QLF
 
 | | MHC CLASS I H-2DB COMPLEXED WITH GLYCOPEPTIDE K3G | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, HUMAN BETA-2-MICROGLOBULIN, ... | | Authors: | Tormo, J, Jones, E.Y. | | Deposit date: | 1999-08-30 | | Release date: | 1999-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Two H-2Db/Glycopeptide Complexes Suggest a Molecular Basis for Ctl Cross-Reactivity
Immunity, 10, 1999
|
|
7ARG
 
 | | Notum in complex with ARUK3002704 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-10-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural Insights into Notum Covalent Inhibition.
J.Med.Chem., 64, 2021
|
|
7B3X
 
 | | Notum complex with ARUK3003748 | | Descriptor: | 6-(m-tolylthio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Fish, P, Jones, E.Y. | | Deposit date: | 2020-12-01 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
7B3I
 
 | | Notum complex with ARUK3003776 | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-chlorophenyl)sulfanyl-1$l^{4},2,7,8-tetrazabicyclo[4.3.0]nona-1(6),2,4,7-tetraen-9-one, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-12-01 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
