1LV5
 
 | | Crystal Structure of the Closed Conformation of Bacillus DNA Polymerase I Fragment Bound to DNA and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*CP*GP*TP*CP*GP*CP*TP*GP*AP*TP*CP*CP*G)-3', 5'-D(*GP*GP*AP*TP*CP*AP*GP*CP*GP*A)-3', ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-05-24 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4QU4
 
 | |
3BZC
 
 | |
3BZK
 
 | |
3Q7E
 
 | |
1L3U
 
 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 11 base pairs of duplex DNA following addition of a dTTP and a dATP residue. | | Descriptor: | 5'-D(*GP*AP*C*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1L5U
 
 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 12 base pairs of duplex DNA following addition of a dTTP, a dATP, and a dCTP residue. | | Descriptor: | 5'-D(*GP*A*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*AP*C)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-08 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1L3S
 
 | | Crystal Structure of Bacillus DNA Polymerase I Fragment complexed to 9 base pairs of duplex DNA. | | Descriptor: | 5'-D(*GP*A*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*G)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1L3V
 
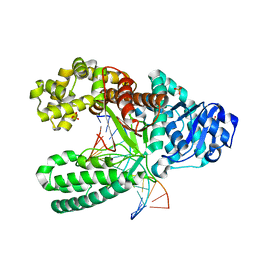 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 15 base pairs of duplex DNA following addition of dTTP, dATP, dCTP, and dGTP residues. | | Descriptor: | 5'-D(*GP*AP*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*AP*CP*GP*TP*C)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1L3T
 
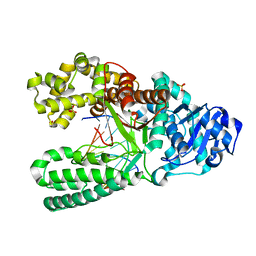 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 10 base pairs of duplex DNA following addition of a single dTTP residue | | Descriptor: | 5'-D(*GP*AP*CP*G*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4YAA
 
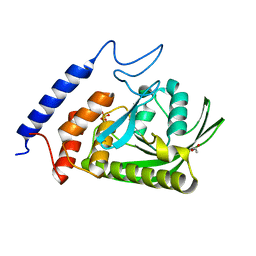 | | YopH W354Y Yersinia enterocolitica PTPase apo form | | Descriptor: | ACETATE ION, SULFATE ION, Tyrosine-protein phosphatase YopH | | Authors: | Moise, G.E, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2015-02-17 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Conservative Tryptophan Mutants of the Protein Tyrosine Phosphatase YopH Exhibit Impaired WPD-Loop Function and Crystallize with Divanadate Esters in Their Active Sites.
Biochemistry, 54, 2015
|
|
5SYR
 
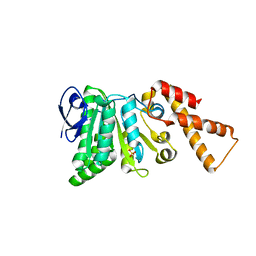 | | Crystal Structure of ATPase delta1-79 Spa47 R350A | | Descriptor: | Probable ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Burgess, J.L, Burgess, R.A, Dickenson, N.E. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of Spa47 Provides Mechanistic Insight into Type III Secretion System ATPase Activation and Shigella Virulence Regulation.
J. Biol. Chem., 291, 2016
|
|
5SWJ
 
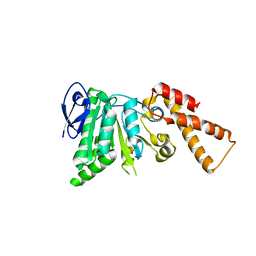 | | Crystal Structure of ATPase delta1-79 Spa47 | | Descriptor: | Probable ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Burgess, J.L, Burgess, R.A, Dickenson, N.E. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural and Biochemical Characterization of Spa47 Provides Mechanistic Insight into Type III Secretion System ATPase Activation and Shigella Virulence Regulation.
J. Biol. Chem., 291, 2016
|
|
5SWL
 
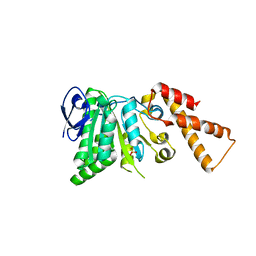 | | Crystal Structure of ATPase delta1-79 Spa47 E188A | | Descriptor: | Probable ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Burgess, J.L, Burgess, R.A, Dickenson, N.E. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Characterization of Spa47 Provides Mechanistic Insight into Type III Secretion System ATPase Activation and Shigella Virulence Regulation.
J. Biol. Chem., 291, 2016
|
|
5SYP
 
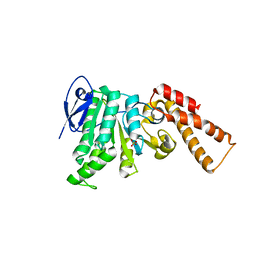 | | Crystal Structure of ATPase delta1-79 Spa47 K165A | | Descriptor: | Probable ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Burgess, J.L, Burgess, R.A, Dickenson, N.E. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Biochemical Characterization of Spa47 Provides Mechanistic Insight into Type III Secretion System ATPase Activation and Shigella Virulence Regulation.
J. Biol. Chem., 291, 2016
|
|
7MPC
 
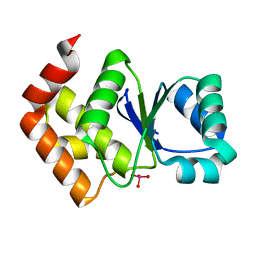 | | Structure of SsoPTP bound to vanadate | | Descriptor: | SsoPTP, VANADATE ION | | Authors: | Pinkston, J.A, Olsen, K.J, Hengge, A.C, Johnson, S.J. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Significant Loop Motions in the SsoPTP Protein Tyrosine Phosphatase Allow for Dual General Acid Functionality.
Biochemistry, 60, 2021
|
|
7MPD
 
 | | Structure of SsoPTP bound to 2-chloroethylsulfonate | | Descriptor: | 2-chloroethane-1-sulfonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SsoPTP | | Authors: | Pinkston, J.A, Olsen, K.J, Hengge, A.C, Johnson, S.J. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Significant Loop Motions in the SsoPTP Protein Tyrosine Phosphatase Allow for Dual General Acid Functionality.
Biochemistry, 60, 2021
|
|
4ZI4
 
 | |
6XEE
 
 | | Crystal Structure of the PTP1B YopH WPD loop Chimera 4 apo form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
6XE8
 
 | | Crystal Structure of the PTP1B YopH WPD loop Chimera 3 apo form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | Authors: | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
6XEA
 
 | | Crystal Structure of the PTP1B YopH WPD loop Chimera 3 bound to vanadate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | Authors: | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
6XEF
 
 | | Crystal structure of the PTP1B YopH WPD loop Chimera 4 bound to vanadate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | Authors: | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
6XED
 
 | | Crystal Structure of the PTP1B YopH WPD loop Chimera 3 bound to tungstate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | Authors: | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
6XEG
 
 | | Crystal structure of the PTP1B YopH WPD loop Chimera 4 bound to tungstate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | Authors: | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
9E9N
 
 | |
