8B6N
 
 | |
8B6P
 
 | |
1K52
 
 | | Monomeric Protein L B1 Domain with a K54G mutation | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K51
 
 | | A G55A Mutation Induces 3D Domain Swapping in the B1 Domain of Protein L from Peptostreptococcus magnus | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
4UQM
 
 | | Crystal structure determination of uracil-DNA N-glycosylase (UNG) from Deinococcus radiodurans in complex with DNA - new insights into the role of the Leucine-loop for damage recognition and repair | | Descriptor: | 5'-D(*CP*CP*TP*AP*TP*CP*CP*AP*AAB*GP*TP*CP*TP*CP*CP*G)-3', 5'-D(*GP*CP*GP*GP*AP*GP*AP*CP*AP*TP*GP*GP*AP*CP*AP*G)-3', CHLORIDE ION, ... | | Authors: | Pedersen, H.L, Johnson, K.A, McVey, C.E, Leiros, I, Moe, E. | | Deposit date: | 2014-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure determination of uracil-DNA N-glycosylase from Deinococcus radiodurans in complex with DNA.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
8B6O
 
 | |
8B6Q
 
 | | X-ray structure of the haloalkane dehalogenase HaloTag7 with an insertion of Calmodulin-M13 fusion at position 154-156 that mimic the structure of CaProLa, an calcium gated protein labeling technology | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase,Calmodulin-1,Haloalkane dehalogenase,Calmodulin-1,M13 peptide | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the haloalkane dehalogenase HaloTag7 with an insertion of Calmodulin-M13 fusion at position 154-156 that mimic the structure of CaProLa, an calcium gated protein labeling technology
To Be Published
|
|
1K6O
 
 | | Crystal Structure of a Ternary SAP-1/SRF/c-fos SRE DNA Complex | | Descriptor: | 5'-D(*CP*AP*CP*AP*GP*GP*AP*TP*GP*TP*CP*CP*AP*TP*AP*TP*TP*AP*GP*GP*AP*CP*A)-3', 5'-D(*TP*GP*TP*CP*CP*TP*AP*AP*TP*AP*TP*GP*GP*AP*CP*AP*TP*CP*CP*TP*GP*TP*G)-3', ETS-domain protein ELK-4, ... | | Authors: | Mo, Y, Ho, W, Johnston, K, Marmorstein, R. | | Deposit date: | 2001-10-16 | | Release date: | 2002-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex.
J.Mol.Biol., 314, 2001
|
|
1F8V
 
 | | THE STRUCTURE OF PARIACOTO VIRUS REVEALS A DODECAHEDRAL CAGE OF DUPLEX RNA | | Descriptor: | CALCIUM ION, MATURE CAPSID PROTEIN BETA, MATURE CAPSID PROTEIN GAMMA, ... | | Authors: | Tang, L, Johnson, K.N, Ball, L.A, Lin, T, Yeager, M, Johnson, J.E. | | Deposit date: | 2000-07-05 | | Release date: | 2000-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of pariacoto virus reveals a dodecahedral cage of duplex RNA.
Nat.Struct.Biol., 8, 2001
|
|
6ENK
 
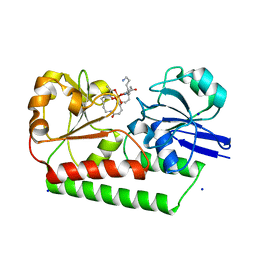 | | The X-ray crystal structure of DesE bound to desferrioxamine B | | Descriptor: | DesE, SODIUM ION, desferrioxamine B | | Authors: | Naismith, J.H, McMahon, S.A, Challis, G.L, Kadi, N, Oke, M, Liu, H, Carter, L.G, Johnson, K.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Desferrioxamine biosynthesis: diverse hydroxamate assembly by substrate-tolerant acyl transferase DesC.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
7PCW
 
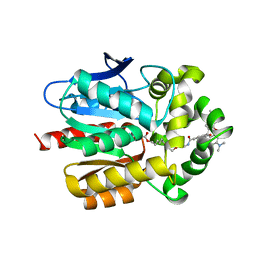 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-M175W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase, [9-[2-carboxy-5-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethyl-azanium | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2021-08-04 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineered HaloTag variants for fluorescence lifetime multiplexing.
Nat.Methods, 19, 2022
|
|
7PCX
 
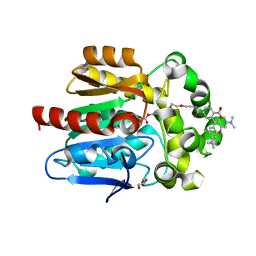 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2021-08-04 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineered HaloTag variants for fluorescence lifetime multiplexing.
Nat.Methods, 19, 2022
|
|
7S4U
 
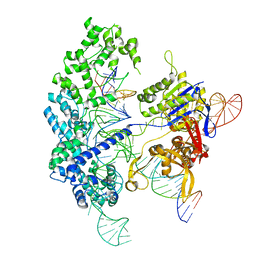 | | Cryo-EM structure of Cas9 in complex with 12-14MM DNA substrate, 5 minute time-point | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand, Target strand, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
7S4V
 
 | | Cas9 bound to 12-14MM DNA, 60 min time-point, kinked conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, NTS, TS, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
7S4X
 
 | | Cas9:gRNA in complex with 18-20MM DNA, 1 minute time-point, kinked active conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
8OVP
 
 | | X-ray structure of the iAspSnFR in complex with L-aspartate | | Descriptor: | ACETATE ION, ASPARTIC ACID, MAGNESIUM ION, ... | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iAspSnFR in complex with L-aspartate
To Be Published
|
|
4AO6
 
 | | Native structure of a novel cold-adapted esterase from an Arctic intertidal metagenomic library | | Descriptor: | ESTERASE | | Authors: | Fu, J, Leiros, H.-K.S, Pascale, D.d, Johnson, K.A, Blencke, H.M, Landfald, B. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional and Structural Studies of a Novel Cold-Adapted Esterase from an Arctic Intertidal Metagenomic Library.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
4AO7
 
 | | Zinc bound structure of a novel cold-adapted esterase from an Arctic intertidal metagenomic library | | Descriptor: | ESTERASE, ZINC ION | | Authors: | Fu, J, Leiros, H.-K.S, Pascale, D.d, Johnson, K.A, Blencke, H.M, Landfald, B. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional and Structural Studies of a Novel Cold-Adapted Esterase from an Arctic Intertidal Metagenomic Library.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
4AO8
 
 | | PEG-bound complex of a novel cold-adapted esterase from an Arctic intertidal metagenomic library | | Descriptor: | DI(HYDROXYETHYL)ETHER, ESTERASE | | Authors: | Fu, J, Leiros, H.-K.S, Pascale, D.d, Johnson, K.A, Blencke, H.M, Landfald, B. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and Structural Studies of a Novel Cold-Adapted Esterase from an Arctic Intertidal Metagenomic Library.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
1LBV
 
 | | Crystal Structure of apo-form (P21) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus | | Descriptor: | fructose 1,6-bisphosphatase/inositol monophosphatase | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBX
 
 | | Crystal Structure of a ternary complex of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus with Calcium ions and D-myo-Inositol-1-Phosphate | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1-PHOSPHATE, fructose 1,6-bisphosphatase/inositol monophosphatase | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBZ
 
 | | Crystal Structure of a complex (P32 crystal form) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus with 3 Calcium ions and Fructose-1,6 bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, fructose 1,6-bisphosphatase/inositol monophosphatase | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBY
 
 | | Crystal Structure of a complex (P32 crystal form) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus with 3 Manganese ions, Fructose-6-Phosphate, and Phosphate ion | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBW
 
 | | Crystal Structure of apo-form (P32) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus | | Descriptor: | fructose 1,6-bisphosphatase/inositol monophosphatase | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
5TZO
 
 | | Computationally Designed Fentanyl Binder - Fen49*-Complex | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase A, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
