1WTU
 
 | | TRANSCRIPTION FACTOR 1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSCRIPTION FACTOR 1 | | Authors: | Jia, X, Grove, A, Ivancic, M, Hsu, V.L, Geiduschek, E.P, Kearns, D.R. | | Deposit date: | 1996-07-29 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Bacillus subtilis phage SPO1-encoded type II DNA-binding protein TF1 in solution.
J.Mol.Biol., 263, 1996
|
|
7YR7
 
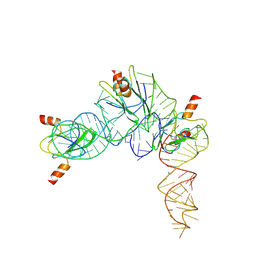 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with three RsmA protein dimers | | Descriptor: | RsmZ RNA (118-MER), Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Liu, L, Ling, X, Yang, X, Wu, Y, Liu, T, Miao, Z, Wei, X, Bujnicki, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
7YR6
 
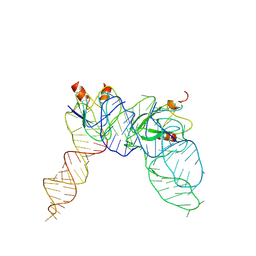 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with two RsmA protein dimers | | Descriptor: | RsmZ RNA, Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Ling, X, Yang, X, Wu, Y, Liu, T, Wei, X, Bujnick, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
6URI
 
 | |
8EPY
 
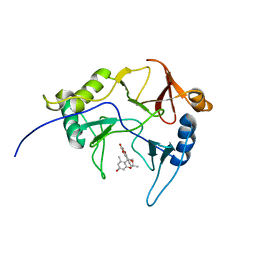 | | The solution structure of abxF in complex with its product (-)-ABX, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX | | Descriptor: | (6R,16R)-3,11,13,15-tetrahydroxy-1,6,9,9-tetramethyl-6,7,9,16-tetrahydro-14H-6,16-epoxyanthra[2,3-e]benzo[b]oxocin-14-one, Glyoxalase | | Authors: | Jia, X, Yan, X, Qu, X, Mobli, M. | | Deposit date: | 2022-10-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of abxF, an enzyme catalyzing the formation of chiral spiroketal of an antibiotics, (-)-ABX.
To Be Published
|
|
8EO9
 
 | | The solution structure of abxF, an enzyme catalyzing the formation of chiral spiroketal of an antibiotics, (-)-ABX | | Descriptor: | Glyoxalase | | Authors: | Jia, X, Yan, X, Mobli, M, Qu, X. | | Deposit date: | 2022-10-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of abxF, an enzyme catalyzing the formation of chiral spiroketal of an antibiotics, (-)-ABX.
To Be Published
|
|
3I17
 
 | |
3CR6
 
 | |
3F8A
 
 | | Crystal Structure of the R132K:R111L:L121E:R59W Mutant of Cellular Retinoic Acid-Binding Protein Type II Complexed with C15-aldehyde (a retinal analog) at 1.95 Angstrom resolution. | | Descriptor: | 1,3,3-trimethyl-2-[(1E,3E)-3-methylpenta-1,3-dien-1-yl]cyclohexene, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular retinoic acid-binding protein 2 | | Authors: | Jia, X, Geiger, J.H. | | Deposit date: | 2008-11-12 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing Wavelength Regulation with an Engineered Rhodopsin Mimic and a C15-Retinal Analogue
Chempluschem, 77, 2012
|
|
3FA6
 
 | |
3F9D
 
 | |
3FEL
 
 | |
3FEP
 
 | | Crystal structure of the R132K:R111L:L121E:R59W-CRABPII mutant complexed with a synthetic ligand (merocyanin) at 2.60 angstrom resolution. | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cellular retinoic acid-binding protein 2 | | Authors: | Jia, X, Geiger, J.H. | | Deposit date: | 2008-11-30 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
3FA9
 
 | |
3FA8
 
 | |
3FEN
 
 | |
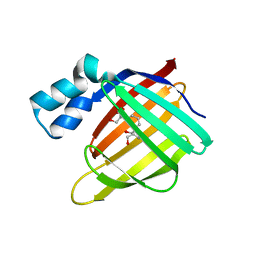
3FA7
 
 | |
3FEK
 
 | |
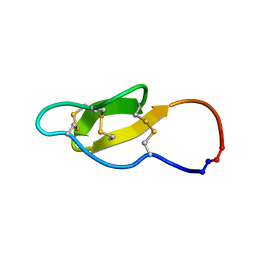
2MH1
 
 | |
4EMZ
 
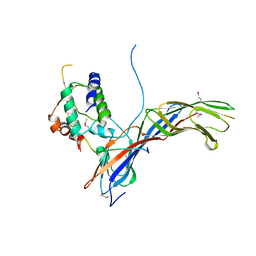 | |
4EN2
 
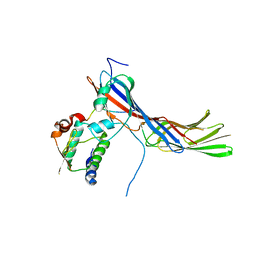 | |
8JON
 
 | | Structure of a synthetic circadian clock protein KaiC mutant of cyanobacteria Synechococcus elongatus PCC 7942 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, ... | | Authors: | Jia, X, Zhang, Q, Li, S, Guo, J. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Adaptation of ancient cyanobacterial clock to the day length ~ 0.95 Ga ago
To Be Published
|
|
4P6Z
 
 | |
5YEP
 
 | | Crystal structure of SO_3166-SO_3165 from Shewanella oneidensis | | Descriptor: | Toxin-antitoxin system antidote Mnt family, Toxin-antitoxin system toxin HepN family | | Authors: | Jia, X, Gao, Z.Q, Zhang, H, Dong, Y.H. | | Deposit date: | 2017-09-19 | | Release date: | 2018-03-28 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function analyses reveal the molecular architecture and neutralization mechanism of a bacterial HEPN-MNT toxin-antitoxin system.
J. Biol. Chem., 293, 2018
|
|
8I8A
 
 | |
