8XFB
 
 | | Cryo-EM structure of partial dimeric WDR11-FAM91A1 complex | | Descriptor: | Protein FAM91A1, WD repeat-containing protein 11 | | Authors: | Jia, G.W, Deng, Q.H, Su, Z.M, Jia, D. | | Deposit date: | 2023-12-13 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The WDR11 complex is a receptor for acidic-cluster-containing cargo proteins.
Cell, 187, 2024
|
|
3ICV
 
 | | Structural Consequences of a Circular Permutation on Lipase B from Candida Antartica | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B | | Authors: | Horton, J.R, Qian, Z, Jia, D, Lutz, S, Cheng, X. | | Deposit date: | 2009-07-18 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation.
J.Mol.Biol., 393, 2009
|
|
3ICW
 
 | | Structure of a Circular Permutation on Lipase B from Candida Antartica with Bound Suicide Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B, PHOSPHATE ION, ... | | Authors: | Horton, J.R, Qian, Z, Jia, D, Lutz, S.A, Cheng, X. | | Deposit date: | 2009-07-18 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation.
J.Mol.Biol., 393, 2009
|
|
6JL7
 
 | |
6JLH
 
 | | Structure of SCGN in complex with a Snap25 peptide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secretagogin, ... | | Authors: | Qin, J, Sun, Q, Jia, D. | | Deposit date: | 2019-03-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and mechanistic insights into secretagogin-mediated exocytosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7D72
 
 | | Cryo-EM structures of human GMPPA/GMPPB complex bound to GDP-Mannose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D73
 
 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (State I) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D74
 
 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (state II) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, Mannose-1-phosphate guanyltransferase beta | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-19 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7XJ2
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C4 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ1
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C2 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ3
 
 | | Structure of human TRPV3 | | Descriptor: | [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, fusion of transient receptor potential cation channel subfamily V member 3 and 3C-GFP | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ0
 
 | | Structure of human TRPV3 in complex with Trpvicin | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
5GVX
 
 | | Structural insight into dephosphorylation by Trehalose 6-phosphate Phosphatase (OtsB2) from Mycobacterium Tuberculosis | | Descriptor: | MAGNESIUM ION, Trehalose-phosphate phosphatase | | Authors: | Shan, S, Min, H, Liu, T, Jiang, D, Rao, Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structural insight into dephosphorylation by trehalose 6-phosphate phosphatase (OtsB2) from Mycobacterium tuberculosis.
FASEB J., 30, 2016
|
|
8GZ1
 
 | | Cryo-EM structure of human NaV1.6/beta1/beta2,apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
8GZ2
 
 | | Cryo-EM structure of human NaV1.6/beta1/beta2-4,9-anhydro-tetrodotoxin | | Descriptor: | (1R,2S,3S,4R,5R,9S,11S,12S,14R)-7-amino-2,4,12-trihydroxy-2-(hydroxymethyl)-10,13,15-trioxa-6,8-diazapentacyclo[7.4.1.1~3,12~.0~5,11~.0~5,14~]pentadec-7-en-8-ium (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
6C1M
 
 | | NavAb NormoPP mutant | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-METHYLGUANIDINE, CHAPSO, ... | | Authors: | Catterall, W.A, Zheng, N, Jiang, D, Gamal El-Din, T.M. | | Deposit date: | 2018-01-04 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Structural basis for gating pore current in periodic paralysis.
Nature, 557, 2018
|
|
6C1P
 
 | | HypoPP mutant | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHAPSO, Ion transport protein, ... | | Authors: | Catterall, W.A, Zheng, N, Jiang, D, Gamal El-Din, T.M. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for gating pore current in periodic paralysis.
Nature, 557, 2018
|
|
6C1K
 
 | | HypoPP mutant with ligand1 | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GUANIDINE, Ion transport protein, ... | | Authors: | Catterall, W.A, Zheng, N, Jiang, D, Gamal El-Din, T.M. | | Deposit date: | 2018-01-04 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for gating pore current in periodic paralysis.
Nature, 557, 2018
|
|
6C1E
 
 | | NavAb NormoPP mutant | | Descriptor: | (3ALPHA,5ALPHA,7ALPHA,8ALPHA,12ALPHA,14BETA,17ALPHA)-3,7,12-TRIHYDROXYCHOL-1-EN-24-AMIDE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5-BETA-24-NOR-CHOLANE-3(ALPHA),7(ALPHA),12(ALPHA)-TRIOL, ... | | Authors: | Catterall, W.A, Zheng, N, Jiang, D, Gamal El-Din, T.M. | | Deposit date: | 2018-01-04 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for gating pore current in periodic paralysis.
Nature, 557, 2018
|
|
4PX7
 
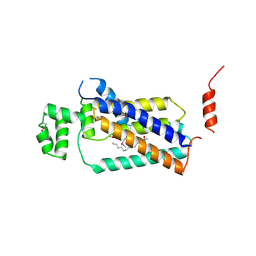 | | Crystal structure of lipid phosphatase E. coli PgpB | | Descriptor: | GLYCEROL, LAURYL DIMETHYLAMINE-N-OXIDE, Phosphatidylglycerophosphatase | | Authors: | Fan, J, Jiang, D, Zhao, Y, Zhang, X.C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of lipid phosphatase Escherichia coli phosphatidylglycerophosphate phosphatase B.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5XKA
 
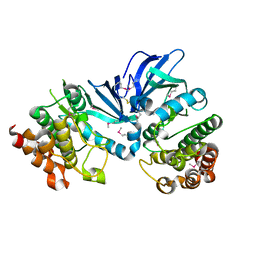 | | Crystal structure of M.tuberculosis PknI kinase domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Yan, Q, Jiang, D, Qian, L, Zhang, Q, Zhang, W, Zhou, W, Mi, K, Guddat, L, Yang, H, Rao, Z. | | Deposit date: | 2017-05-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
5X7F
 
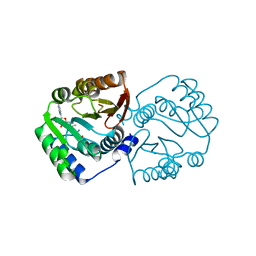 | |
