7Y0J
 
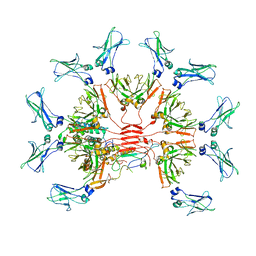 | |
7Y0H
 
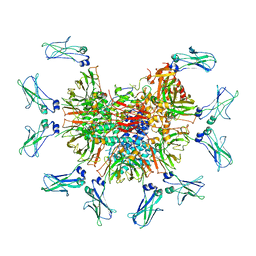 | |
3D9H
 
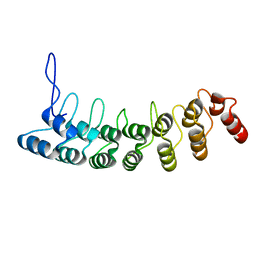 | | Crystal Structure of the Splice Variant of Human ASB9 (hASB9-2), an Ankyrin Repeat Protein | | Descriptor: | cDNA FLJ77766, highly similar to Homo sapiens ankyrin repeat and SOCS box-containing 9 (ASB9), transcript variant 2, ... | | Authors: | Fei, X, Gu, X, Fan, S, Zhang, C, Ji, C. | | Deposit date: | 2008-05-27 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Splice Variant of Human ASB9 (hASB9-2), an Ankyrin Repeat Protein
To be published
|
|
6VX6
 
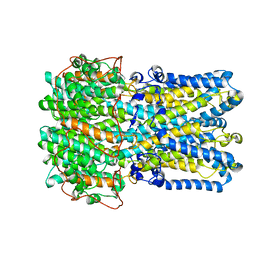 | | bestrophin-2 Ca2+-bound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX5
 
 | | bestrophin-2 Ca2+- unbound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX8
 
 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX9
 
 | | bestrophin-2 Ca2+- unbound state 1 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX7
 
 | | bestrophin-2 Ca2+-bound state (5 mM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2A7R
 
 | | Crystal structure of human Guanosine Monophosphate reductase 2 (GMPR2) | | Descriptor: | GMP reductase 2, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION | | Authors: | Li, J, Wei, Z, Zheng, M, Gu, X, Deng, Y, Qiu, R, Chen, F, Ji, C, Gong, W, Xie, Y, Mao, Y. | | Deposit date: | 2005-07-05 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Human Guanosine Monophosphate Reductase 2 (GMPR2) in Complex with GMP
J.Mol.Biol., 355, 2006
|
|
4O8C
 
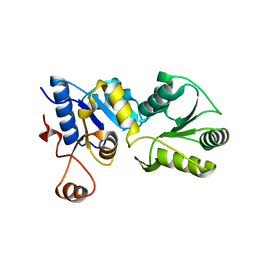 | | Structure of the H170Y mutant of thermostable p-nitrophenylphosphatase from Bacillus Stearothermophilus | | Descriptor: | MAGNESIUM ION, SULFATE ION, Thermostable NPPase | | Authors: | Shen, T, Guo, Z, Wang, F, Gong, W, Ji, C. | | Deposit date: | 2013-12-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a His170Tyr mutant of thermostable pNPPase from Geobacillus stearothermophilus.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3UTN
 
 | | Crystal structure of Tum1 protein from Saccharomyces cerevisiae | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Thiosulfate sulfurtransferase TUM1 | | Authors: | Qiu, R, Wang, F, Liu, M, Ji, C, Gong, W. | | Deposit date: | 2011-11-26 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Tum1 protein from the yeast Saccharomyces cerevisiae.
Protein Pept.Lett., 19, 2012
|
|
4HCC
 
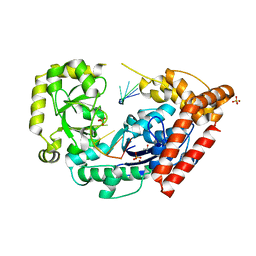 | | The zinc ion bound form of crystal structure of E.coli ExoI-ssDNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Exodeoxyribonuclease I, ISOPROPYL ALCOHOL, ... | | Authors: | Qiu, R, Wei, J, Lou, T, Liu, M, Ji, C, Gong, W. | | Deposit date: | 2012-09-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The structures of Escherichia coli exonuclease I in complex with the single strand DNA
To be published
|
|
4H86
 
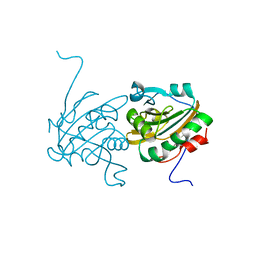 | | Crystal structure of Ahp1 from Saccharomyces cerevisiae in reduced form | | Descriptor: | Peroxiredoxin type-2 | | Authors: | Liu, M, Wang, F, Qiu, R, Wu, T, Gu, S, Tang, R, Ji, C. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure of Ahp1 from Saccharomyces cerevisiae in reduced form
To be Published
|
|
4HCB
 
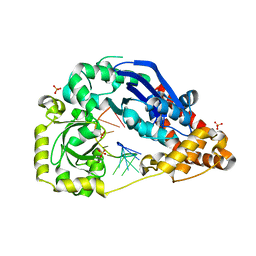 | | The metal-free form of crystal structure of E.coli ExoI-ssDNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Exodeoxyribonuclease I, GLYCEROL, ... | | Authors: | Qiu, R, Wei, J, Lou, T, Liu, M, Ji, C, Gong, W. | | Deposit date: | 2012-09-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of Escherichia coli exonuclease I in complex with the single strand DNA
To be published
|
|
4KN8
 
 | | Crystal structure of Bs-TpNPPase | | Descriptor: | Thermostable NPPase | | Authors: | Guo, Z, Wang, F, Huang, J, Gong, W, Ji, C. | | Deposit date: | 2013-05-09 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal Structure of Thermostable p-nitrophenylphosphatase from Bacillus Stearothermophilus (Bs-TpNPPase)
PROTEIN PEPT.LETT., 21, 2014
|
|
4IFT
 
 | | Crystal structure of double mutant thermostable NPPase from Geobacillus stearothermophilus | | Descriptor: | Thermostable NPPase | | Authors: | Guo, Z, Huang, J, Wang, F, Qiu, R, Wang, Y, Ji, C. | | Deposit date: | 2012-12-15 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Crystal structure of thermostable NPPase from Geobacillus stearothermophilus
To be Published
|
|
4IG4
 
 | | Crystal structure of single mutant thermostable NPPase (N86S) from Geobacillus stearothermophilus | | Descriptor: | Thermostable NPPase | | Authors: | Guo, Z, Wang, F, Huang, J, Qiu, R, Yang, Z, Wang, Y, Gong, W, Ji, C. | | Deposit date: | 2012-12-16 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structure of thermostable NPPase from Geobacillus stearothermophilus
To be Published
|
|
4S2S
 
 | | Crystal Structure of Fab fragment of monoclonal antibody RoAb13 | | Descriptor: | RoAb13 Fab Heavy chain, RoAb13 Fab Light chain | | Authors: | Chain, B, Arnold, J, Akthar, S, Noursadeghi, M, Lapp, T, Ji, C, Naider, D, Zhang, Y, Govada, L, Saridakis, E, Chayen, N.E. | | Deposit date: | 2015-01-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Linear Epitope in the N-Terminal Domain of CCR5 and Its Interaction with Antibody.
Plos One, 10, 2015
|
|
7Y09
 
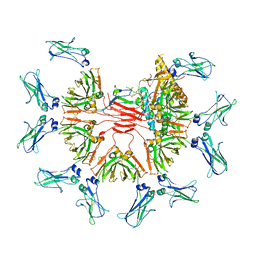 | | Cryo-EM structure of human IgM-Fc in complex with the J chain and the DBL domain of DBLMSP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Shen, H, Ji, C, Xiao, J. | | Deposit date: | 2022-06-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Plasmodium falciparum has evolved multiple mechanisms to hijack human immunoglobulin M.
Nat Commun, 14, 2023
|
|
7YG2
 
 | | Cryo-EM structure of human IgM-Fc in complex with the J chain and the DBL domain of DBLMSP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DBLMSP2, ... | | Authors: | Shen, H, Ji, C, Xiao, J. | | Deposit date: | 2022-07-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Plasmodium falciparum has evolved multiple mechanisms to hijack human immunoglobulin M.
Nat Commun, 14, 2023
|
|
