7CA3
 
 | | Cryo-EM structure of human GABA(B) receptor bound to the positive allosteric modulator rac-BHFF | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, CHOLESTEROL, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | Deposit date: | 2020-06-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
7CUM
 
 | | Cryo-EM structure of human GABA(B) receptor bound to the antagonist CGP54626 | | Descriptor: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, CHOLESTEROL, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | Deposit date: | 2020-08-23 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
7CA5
 
 | | Cryo-EM structure of human GABA(B) receptor in apo state | | Descriptor: | Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | Deposit date: | 2020-06-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
7EWP
 
 | | Cryo-EM structure of human GPR158 in complex with RGS7-Gbeta5 in a 2:1:1 ratio | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Probable G-protein coupled receptor 158, Regulator of G-protein signaling 7 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Cho, Y. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Nat Commun, 12, 2021
|
|
7EWL
 
 | | cryo-EM structure of apo GPR158 | | Descriptor: | Probable G-protein coupled receptor 158 | | Authors: | Jeong, E, Kim, Y, Jeong, J, Cho, Y. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Nat Commun, 12, 2021
|
|
7EWR
 
 | | Cryo-EM structure of human GPR158 in complex with RGS7-Gbeta5 in a 2:2:2 ratio | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Probable G-protein coupled receptor 158, Regulator of G-protein signaling 7 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Cho, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Nat Commun, 12, 2021
|
|
1UA4
 
 | | Crystal Structure of an ADP-dependent Glucokinase from Pyrococcus furiosus | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-dependent glucokinase, alpha-D-glucopyranose, ... | | Authors: | Ito, S, Jeong, J.J, Yoshioka, I, Koga, S, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2003-02-27 | | Release date: | 2004-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an ADP-dependent glucokinase from Pyrococcus furiosus: implications for a sugar-induced conformational change in ADP-dependent kinase
J.Mol.Biol., 331, 2003
|
|
4D2K
 
 | |
5G2G
 
 | |
4L7M
 
 | |
7YQ8
 
 | | Cryo-EM structure of human topoisomerase II beta in complex with DNA and etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, 50-mer DNA, DNA topoisomerase 2-beta, ... | | Authors: | Naganuma, M, Ehara, H, Kim, D, Nakagawa, R, Cong, A, Bu, H, Jeong, J, Jang, J, Schellenberg, M.J, Bunch, H, Sekine, S. | | Deposit date: | 2022-08-05 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ERK2-topoisomerase II regulatory axis is important for gene activation in immediate early genes.
Nat Commun, 14, 2023
|
|
8I7Q
 
 | |
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | Descriptor: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | Authors: | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | Deposit date: | 2020-07-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
5AI1
 
 | | Crystal structure of ketosteroid isomerase containing Y32F, D40N, Y57F and Y119F mutations in the equilenin-bound form | | Descriptor: | EQUILENIN, KETOSTEROID ISOMERASE | | Authors: | Cha, H.J, Jeong, J.H, Kim, Y.G. | | Deposit date: | 2015-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Contribution of a Low-Barrier Hydrogen Bond to Catalysis is not Significant in Ketosteroid Isomerase.
Mol.Cells, 38, 2015
|
|
5XPC
 
 | | Crystal Structure of Drep4 CIDE domain | | Descriptor: | DNAation factor-related protein 4, GLYCEROL | | Authors: | Park, H.H, Jeong, J.H. | | Deposit date: | 2017-06-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | CIDE domains form functionally important higher-order assemblies for DNA fragmentation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7C3K
 
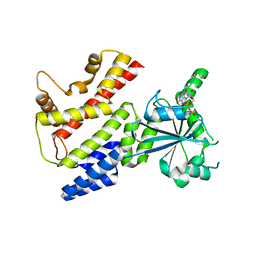 | | Crystal Structure of mIRGB10 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Immunity-related GTPase family member b10 | | Authors: | Ha, H.J, Jeong, J.H, Kim, Y.G, Park, H.H. | | Deposit date: | 2020-05-12 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of IRGB10 oligomerization and membrane association for pathogen membrane disruption.
Commun Biol, 4, 2021
|
|
5XPB
 
 | |
